Summary information and primary citation
- PDB-id
- 1iv6; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- NMR
- Summary
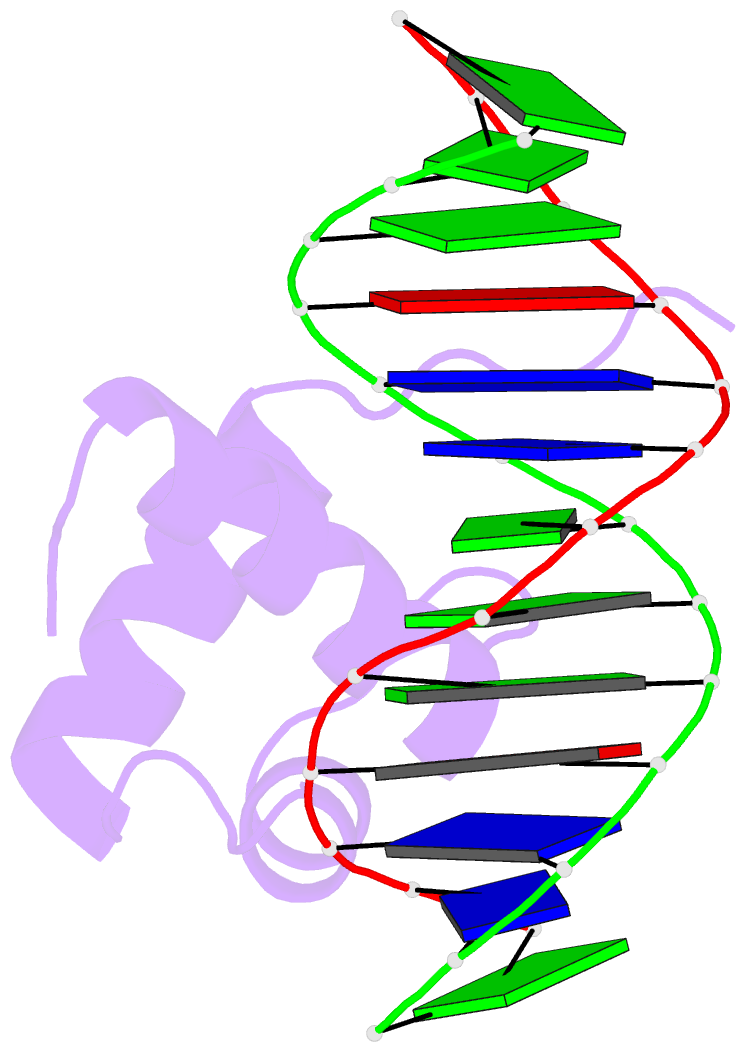
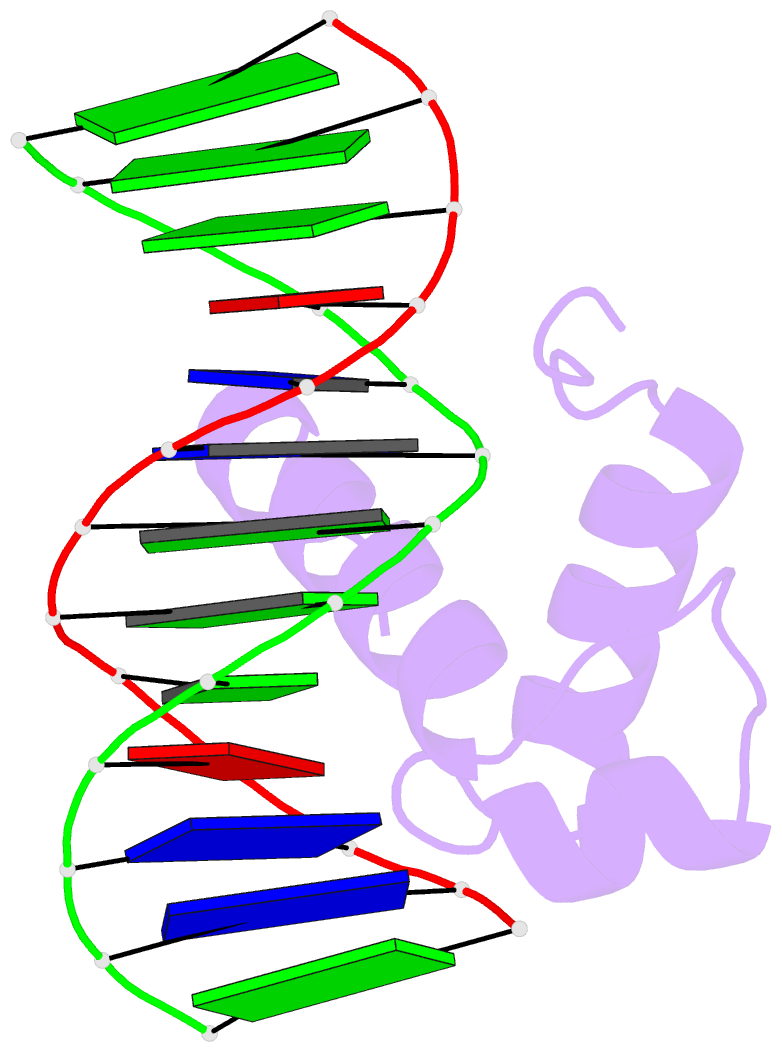
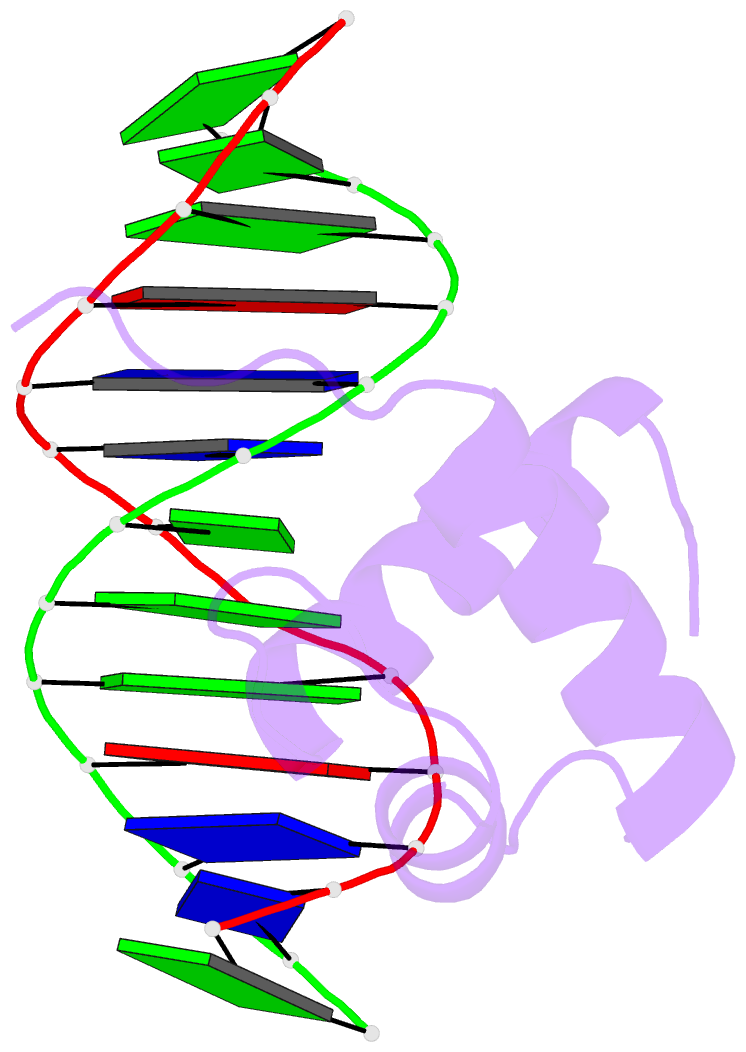
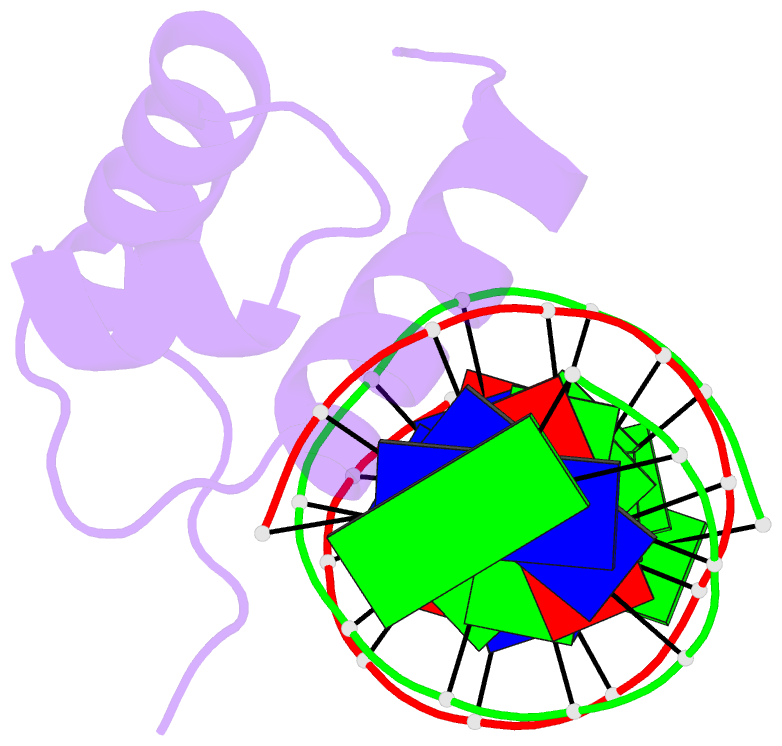
- Solution structure of the DNA complex of human trf1
- Reference
- Nishikawa T, Okamura H, Nagadoi A, Konig P, Rhodes D, Nishimura Y (2001): "Solution structure of a telomeric DNA complex of human TRF1." Structure, 9, 1237-1251. doi: 10.1016/S0969-2126(01)00688-8.
- Abstract
- Background: Mammalian telomeres consist of long tandem arrays of double-stranded TTAGGG sequence motif packaged by TRF1 and TRF2. In contrast to the DNA binding domain of c-Myb, which consists of three imperfect tandem repeats, DNA binding domains of both TRF1 and TRF2 contain only a single Myb repeat. In a DNA complex of c-Myb, both the second and third repeats are closely packed in the major groove of DNA and recognize a specific base sequence cooperatively.
Results: The structure of the DNA binding domain of human TRF1 bound to telomeric DNA has been determined by NMR. It consists of three helices, whose architecture is very close to that of three repeats of the c-Myb DNA binding domain. Only the single Myb domain of TRF1 is sufficient for the sequence-specific recognition. The third helix of TRF1 recognizes the TAGGG part in the major groove, and the N-terminal arm interacts with the TT part in the minor groove.
Conclusions: The DNA binding domain of TRF1 can specifically and fully recognize the AGGGTT sequence. It is likely that, in the dimer of TRF1, two DNA binding domains can bind independently in tandem arrays to two binding sites of telomeric DNA that is composed of the repeated AGGGTT motif. Although TRF2 plays an important role in the t loop formation that protects the ends of telomeres, it is likely that the binding mode of TRF2 to double-stranded telomeric DNA is almost identical to that of TRF1.