Summary information and primary citation
- PDB-id
- 1j1u; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ligase-RNA
- Method
- X-ray (1.95 Å)
- Summary
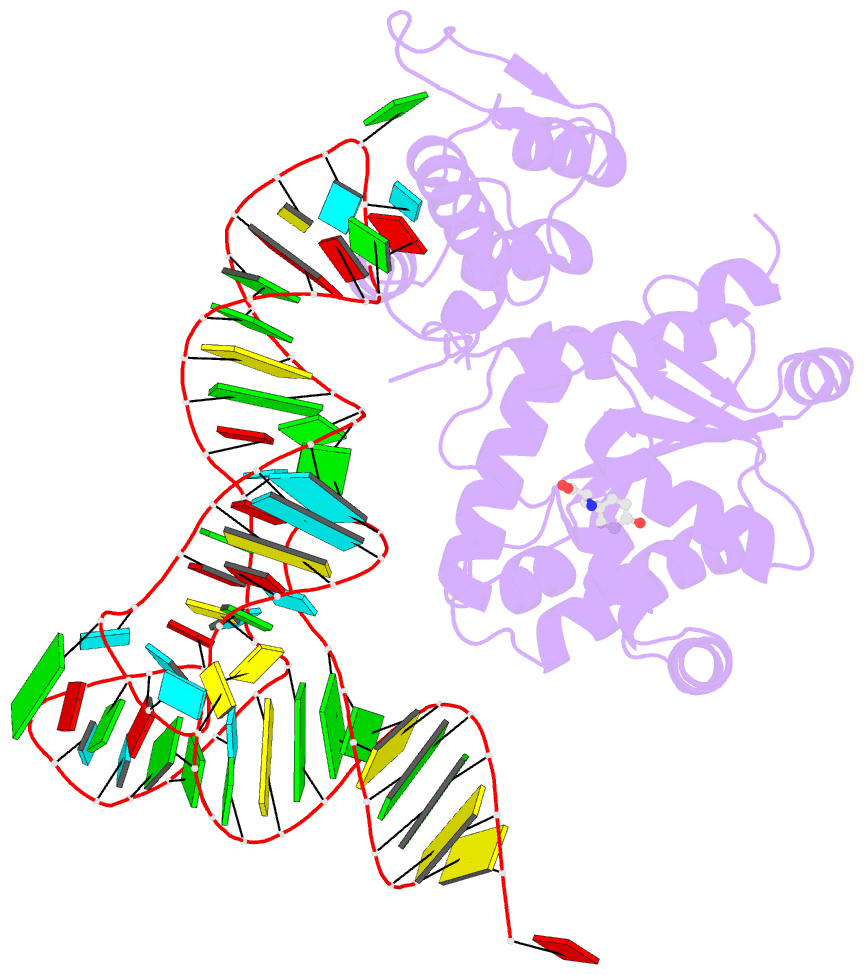
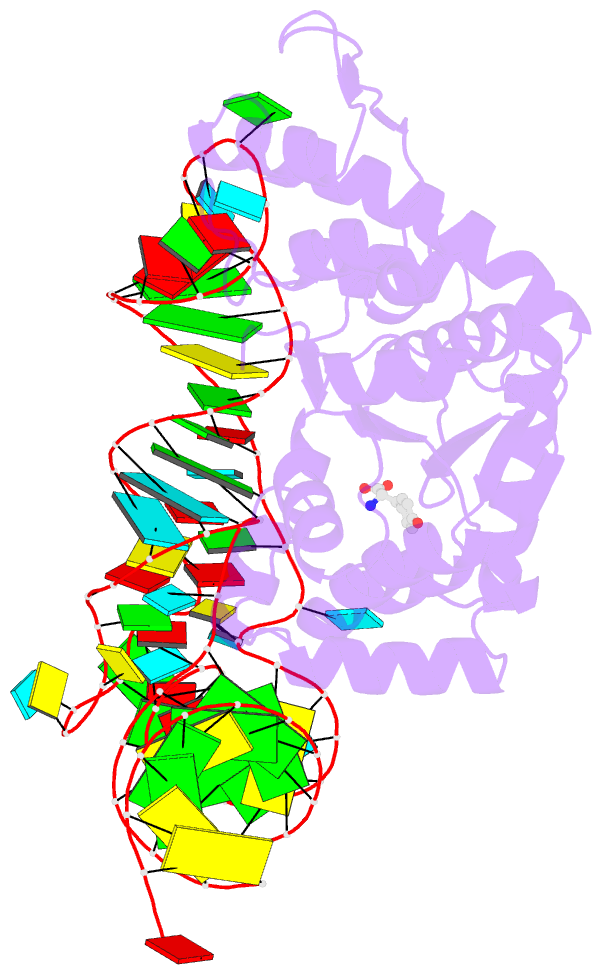
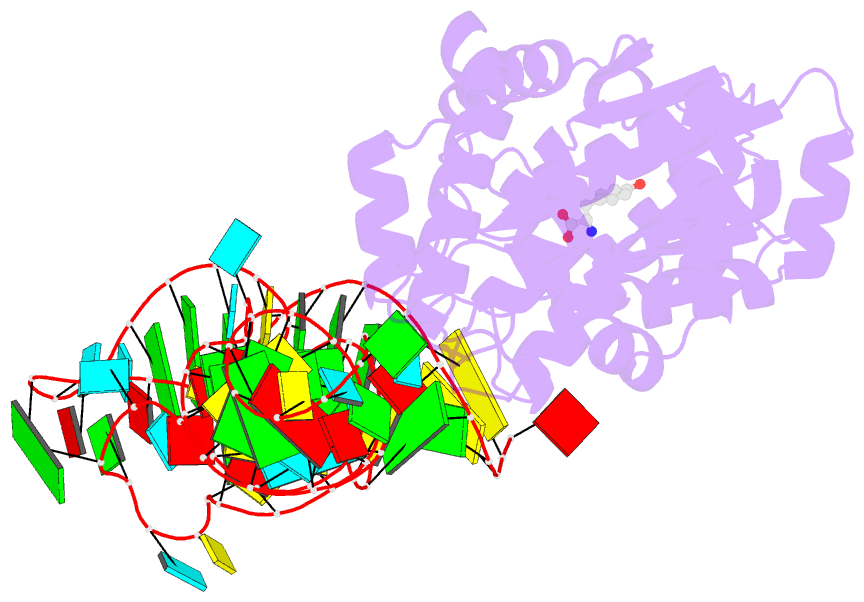
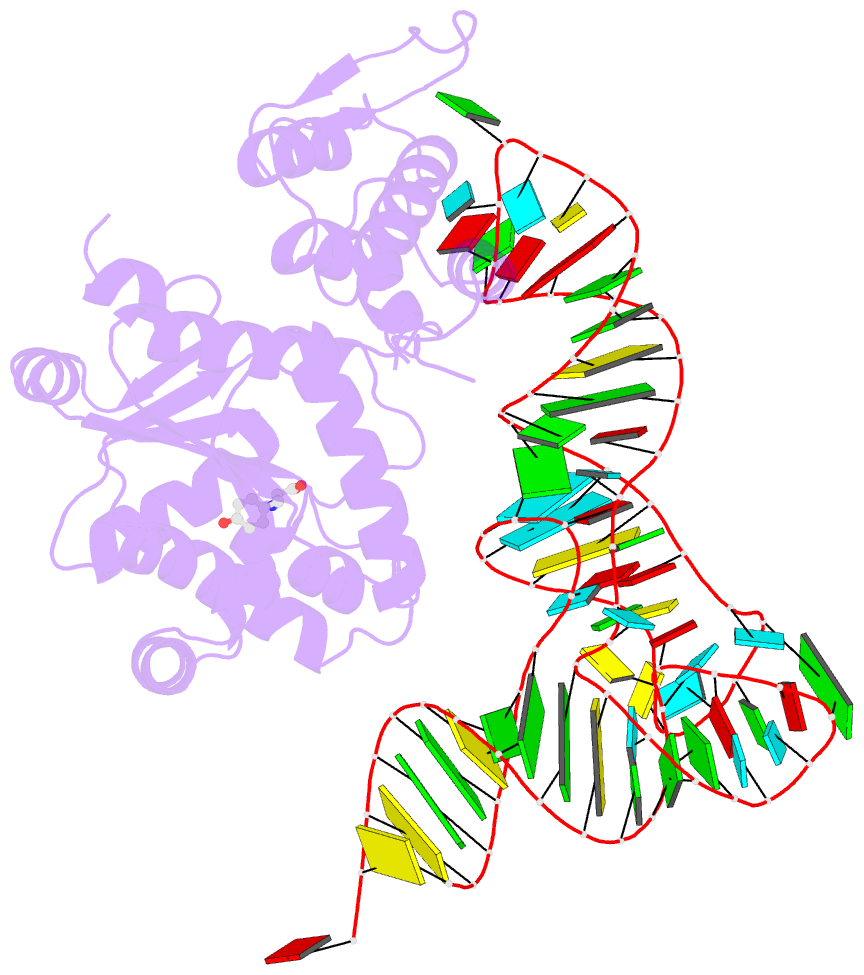
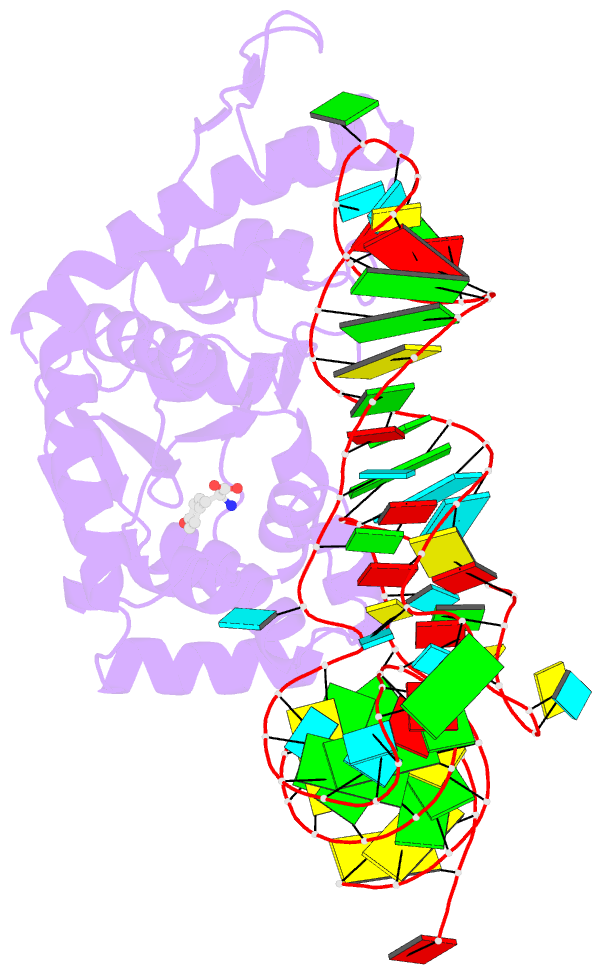
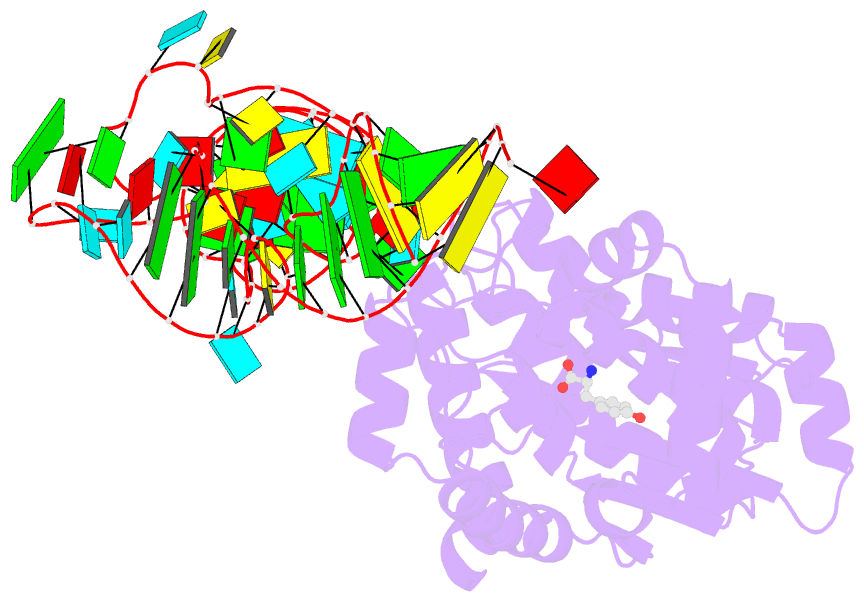
- Crystal structure of archaeal tyrosyl-trna synthetase complexed with trna(tyr) and l-tyrosine
- Reference
- Kobayashi T, Nureki O, Ishitani R, Yaremchuk A, Tukalo M, Cusack S, Sakamoto K, Yokoyama S (2003): "Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion." NAT.STRUCT.BIOL., 10, 425-432. doi: 10.1038/nsb934.
- Abstract
- The archaeal/eukaryotic tyrosyl-tRNA synthetase (TyrRS)-tRNA(Tyr) pairs do not cross-react with their bacterial counterparts. This 'orthogonal' condition is essential for using the archaeal pair to expand the bacterial genetic code. In this study, the structure of the Methanococcus jannaschii TyrRS-tRNA(Tyr)-L-tyrosine complex, solved at a resolution of 1.95 A, reveals that this archaeal TyrRS strictly recognizes the C1-G72 base pair, whereas the bacterial TyrRS recognizes the G1-C72 in a different manner using different residues. These diverse tRNA recognition modes form the basis for the orthogonality. The common tRNA(Tyr) identity determinants (the discriminator, A73 and the anticodon residues) are also recognized in manners different from those of the bacterial TyrRS. Based on this finding, we created a mutant TyrRS that aminoacylates the amber suppressor tRNA with C34 65 times more efficiently than does the wild-type enzyme.