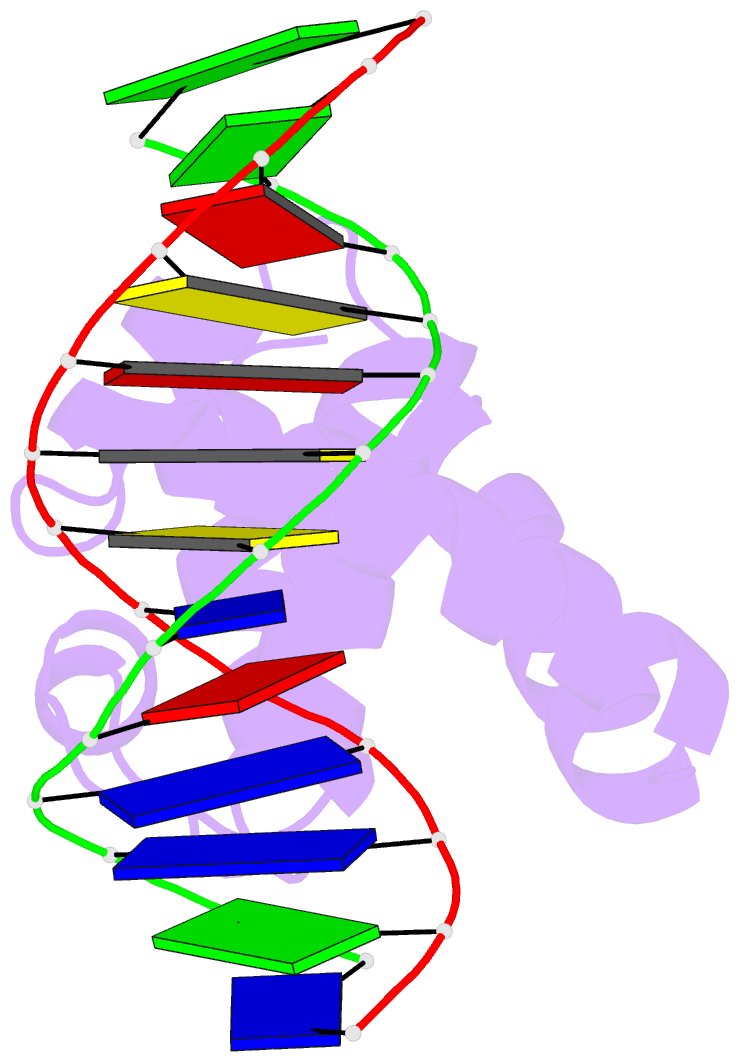
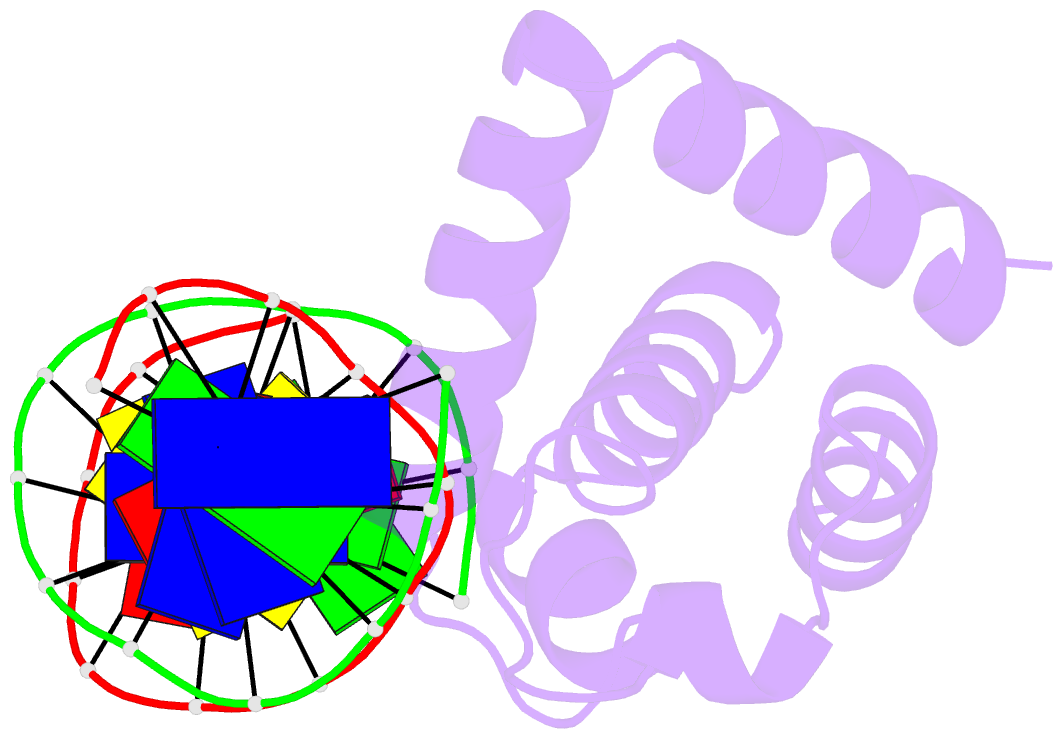
Summary information and primary citation
- PDB-id
- 1j1v; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication-DNA
- Method
- X-ray (2.1 Å)
- Summary
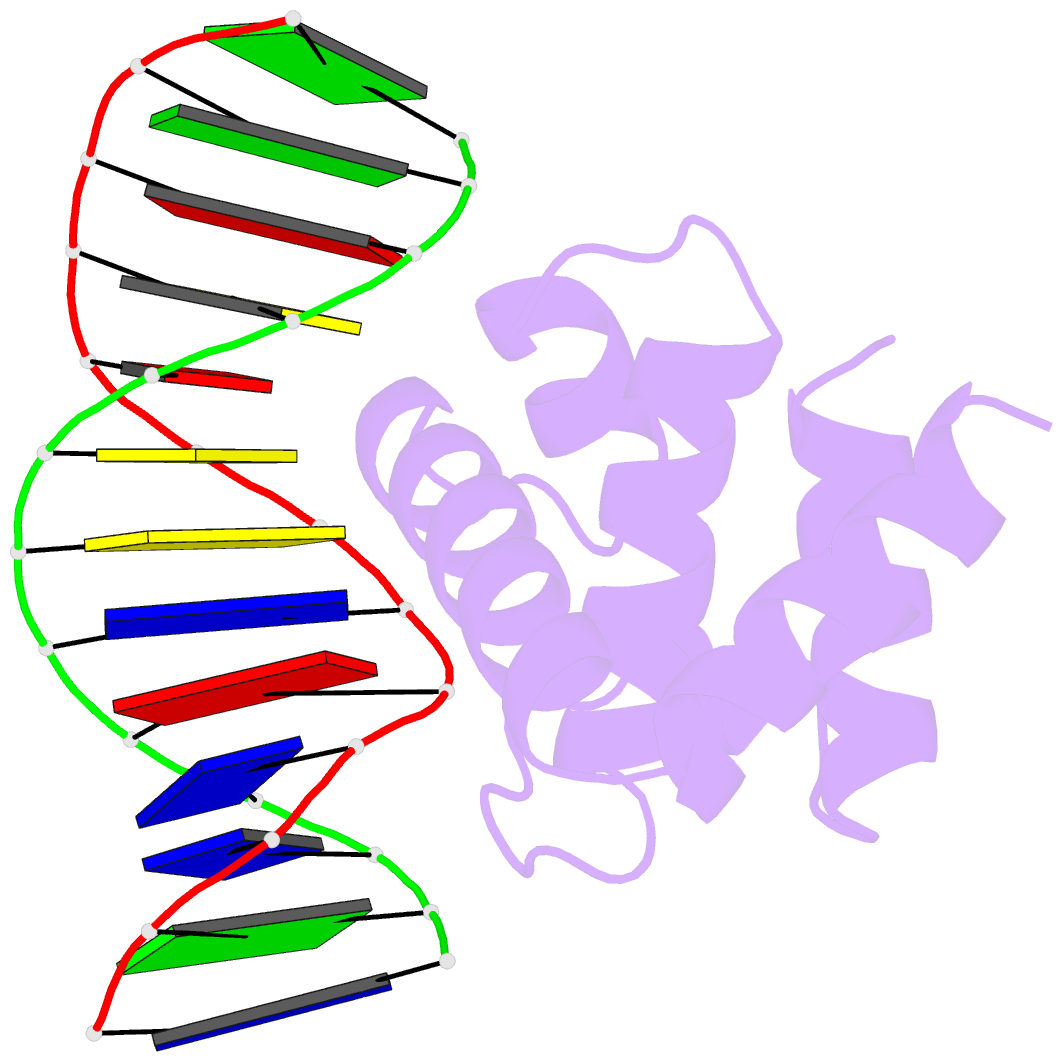
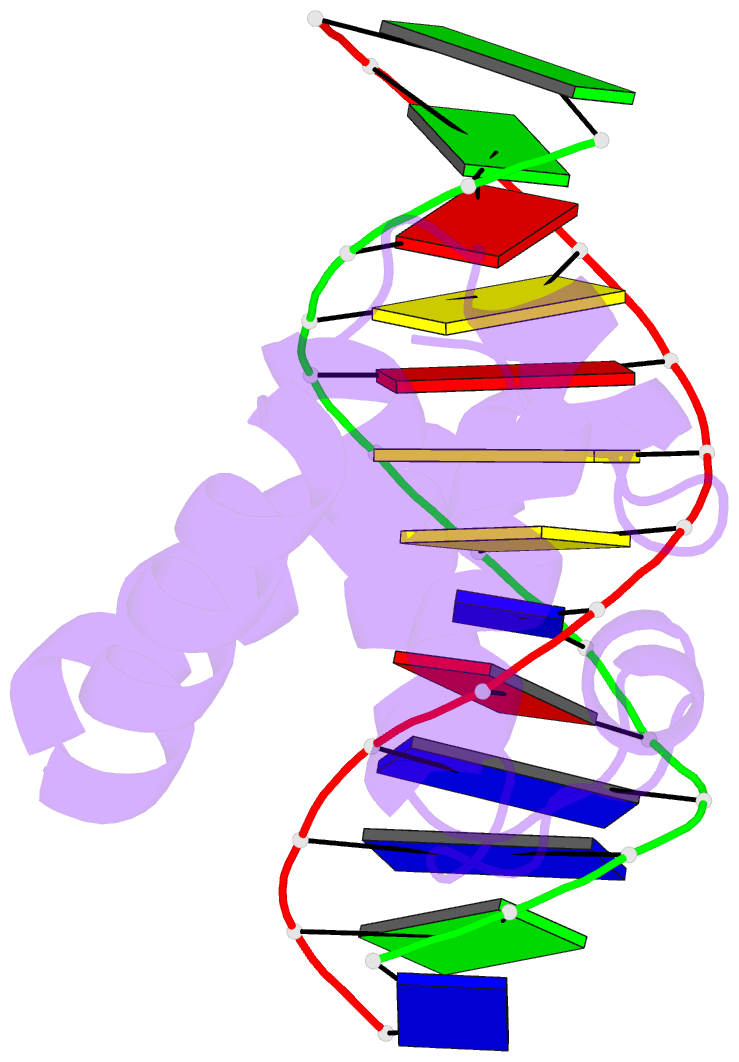
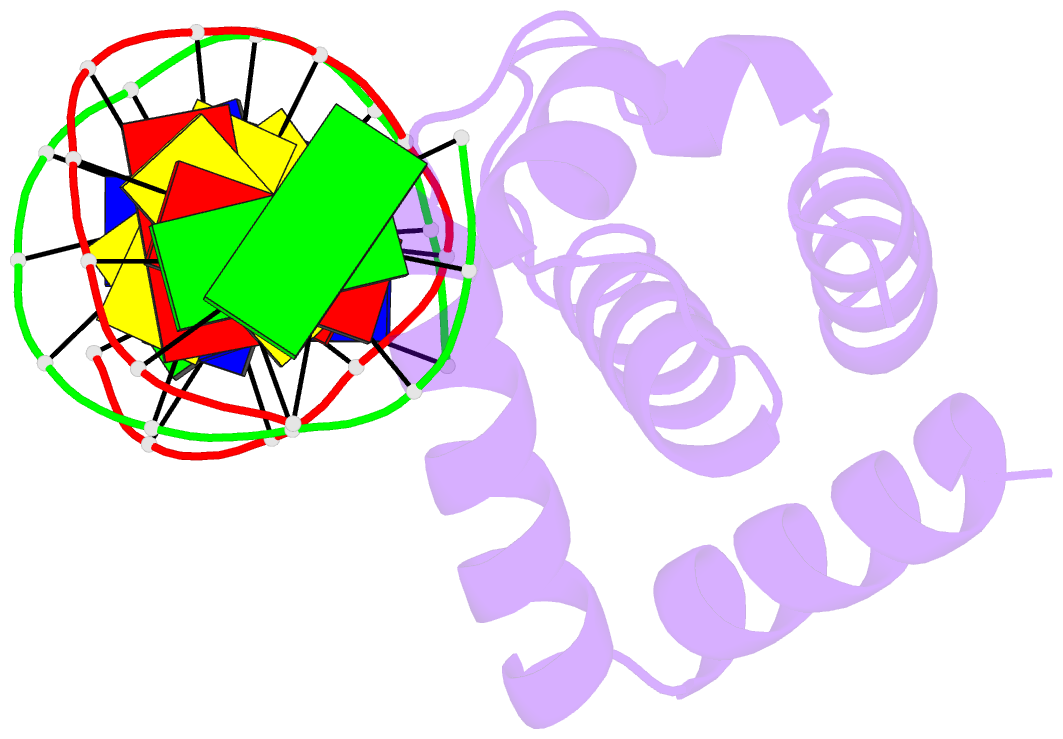
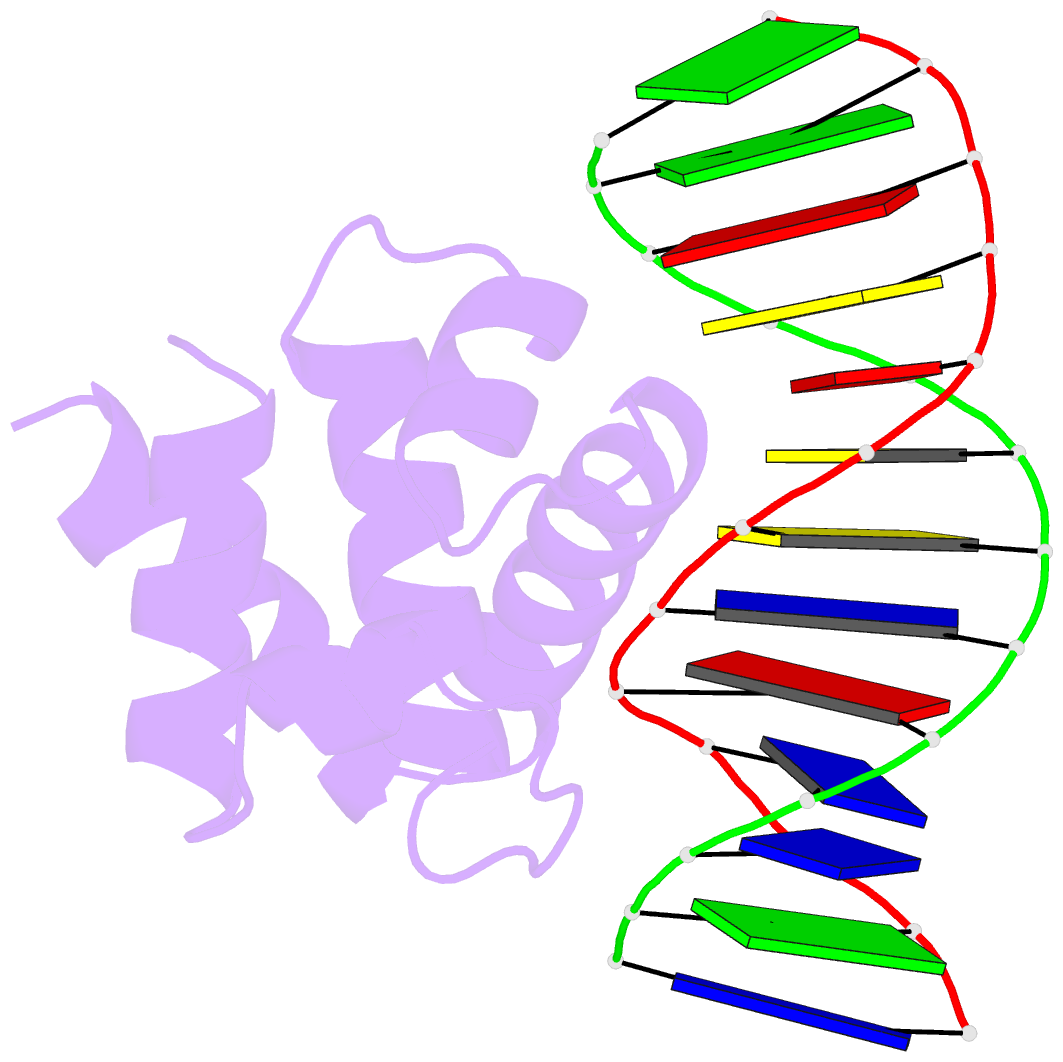
- Crystal structure of dnaa domainiv complexed with dnaabox DNA
- Reference
- Fujikawa N, Kurumizaka H, Nureki O, Terada T, Shirouzu M, Katayama T, Yokoyama S (2003): "Structural basis of replication origin recognition by the DnaA protein." NUCLEIC ACIDS RES., 31, 2077-2086. doi: 10.1093/nar/gkg309.
- Abstract
- Escherichia coli DnaA binds to 9 bp sequences (DnaA boxes) in the replication origin, oriC, to form a complex initiating chromosomal DNA replication. In the present study, we determined the crystal structure of its DNA-binding domain (domain IV) complexed with a DnaA box at 2.1 A resolution. DnaA domain IV contains a helix-turn-helix motif for DNA binding. One helix and a loop of the helix- turn-helix motif are inserted into the major groove and 5 bp (3' two-thirds of the DnaA box sequence) are recognized through base-specific hydrogen bonds and van der Waals contacts with the C5-methyl groups of thymines. In the minor groove, Arg399, located in the loop adjacent to the motif, recognizes three more base pairs (5' one-third of the DnaA box sequence) by base-specific hydrogen bonds. DNA bending by approximately 28 degrees was also observed in the complex. These base-specific interactions explain how DnaA exhibits higher affinity for the strong DnaA boxes (R1, R2 and R4) than the weak DnaA boxes (R3 and M) in the replication origin.