Summary information and primary citation
- PDB-id
- 1j2b; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-RNA
- Method
- X-ray (3.3 Å)
- Summary
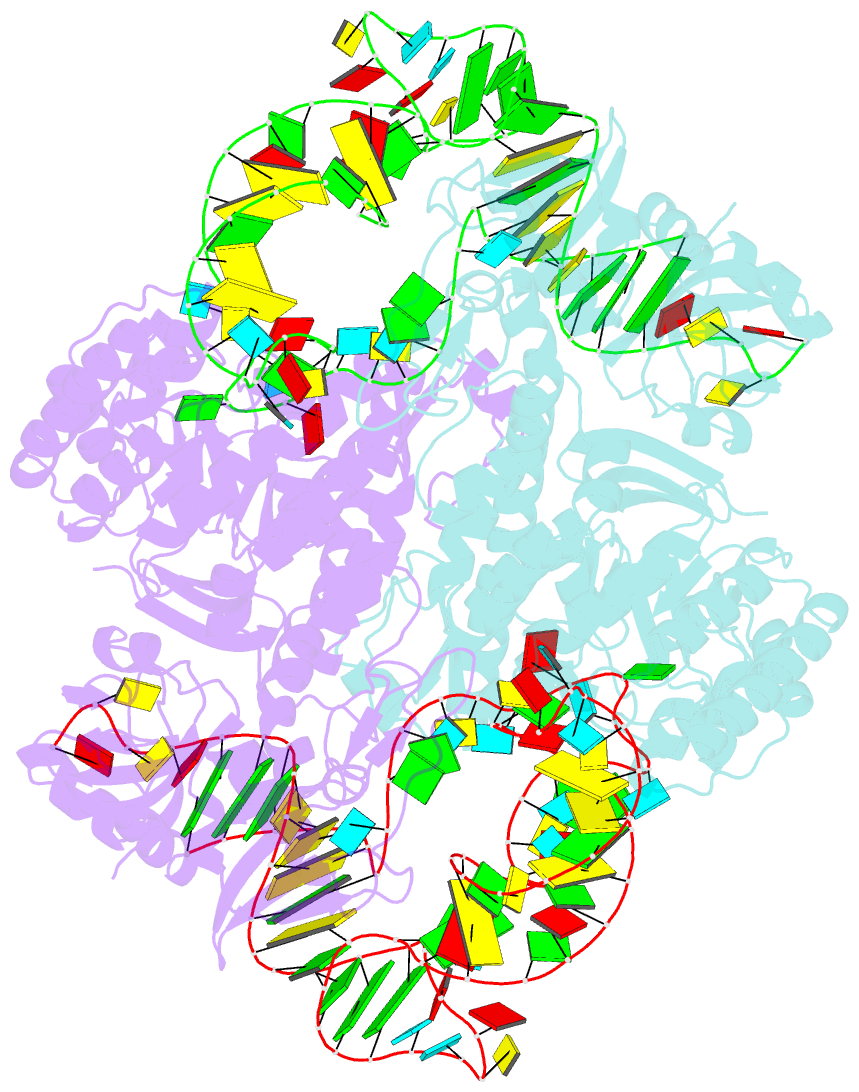
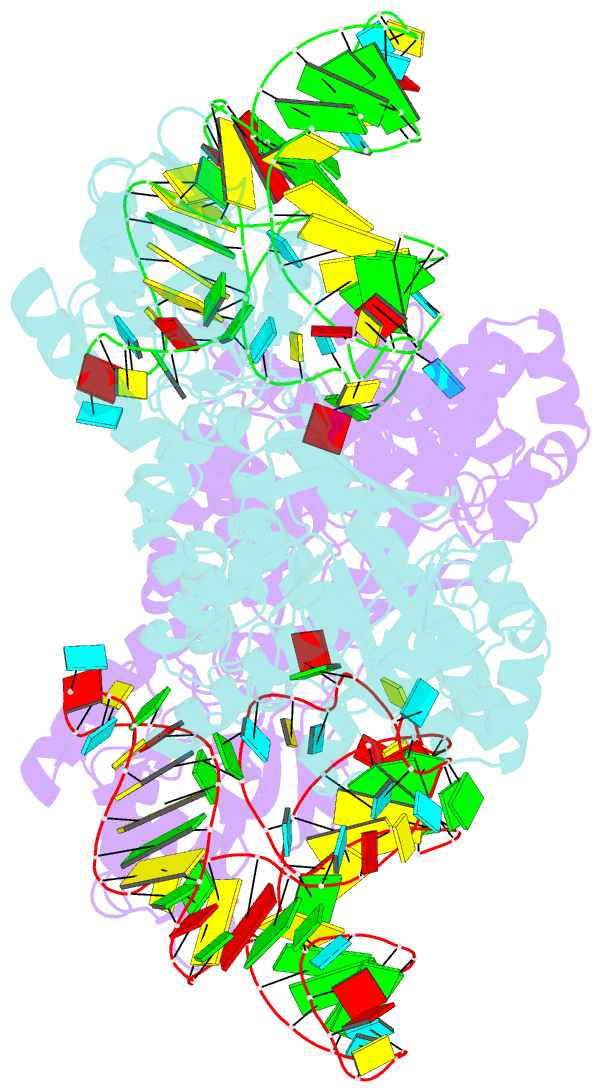
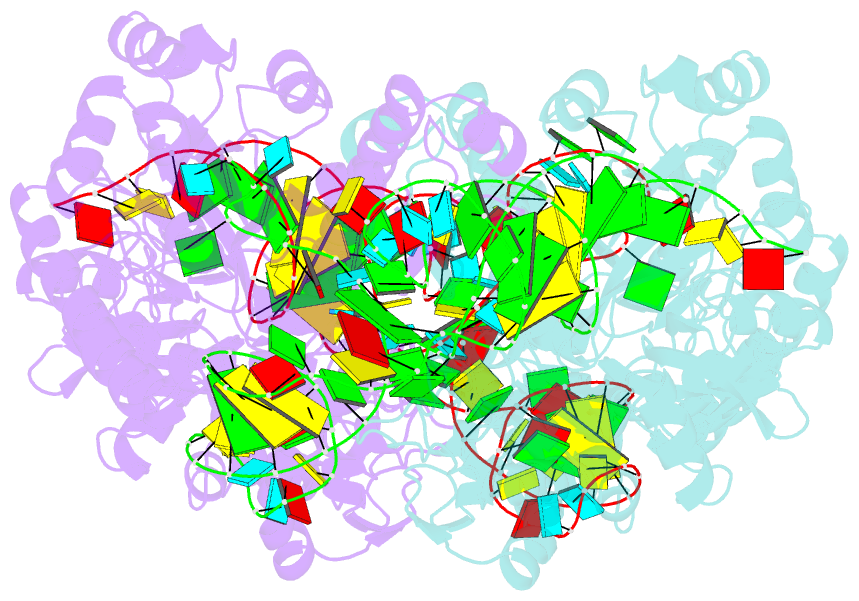
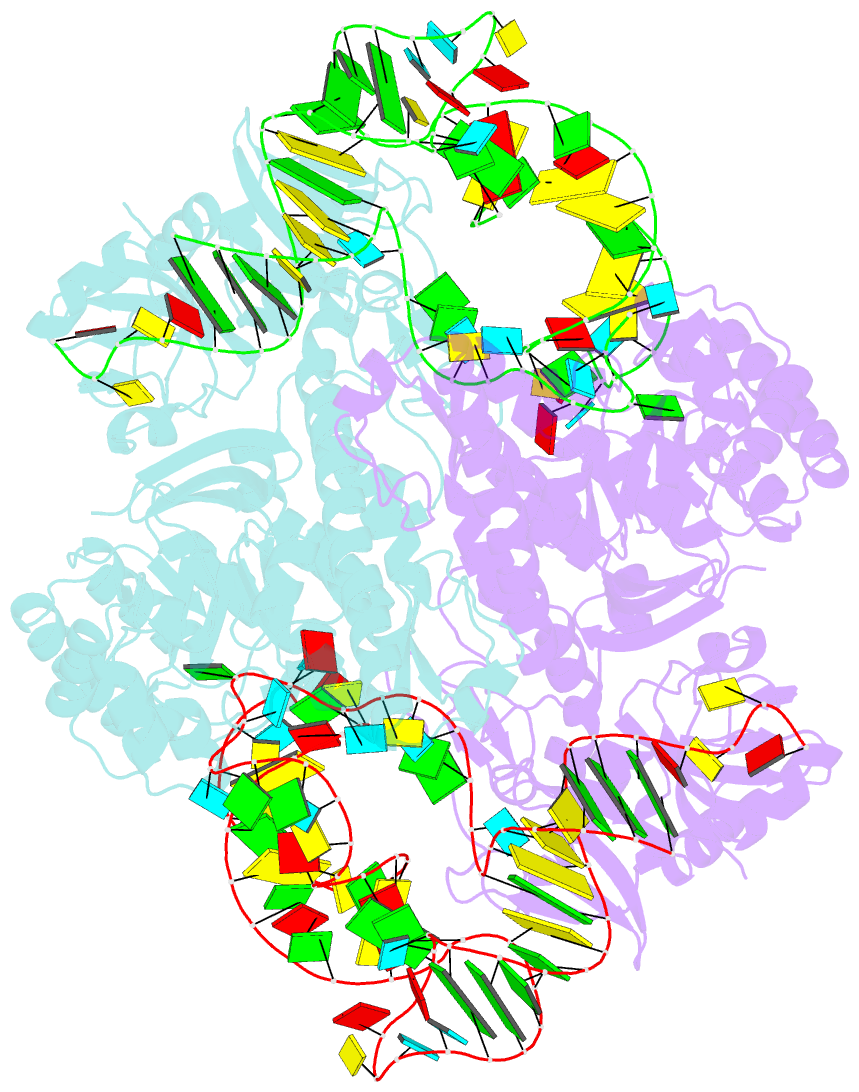
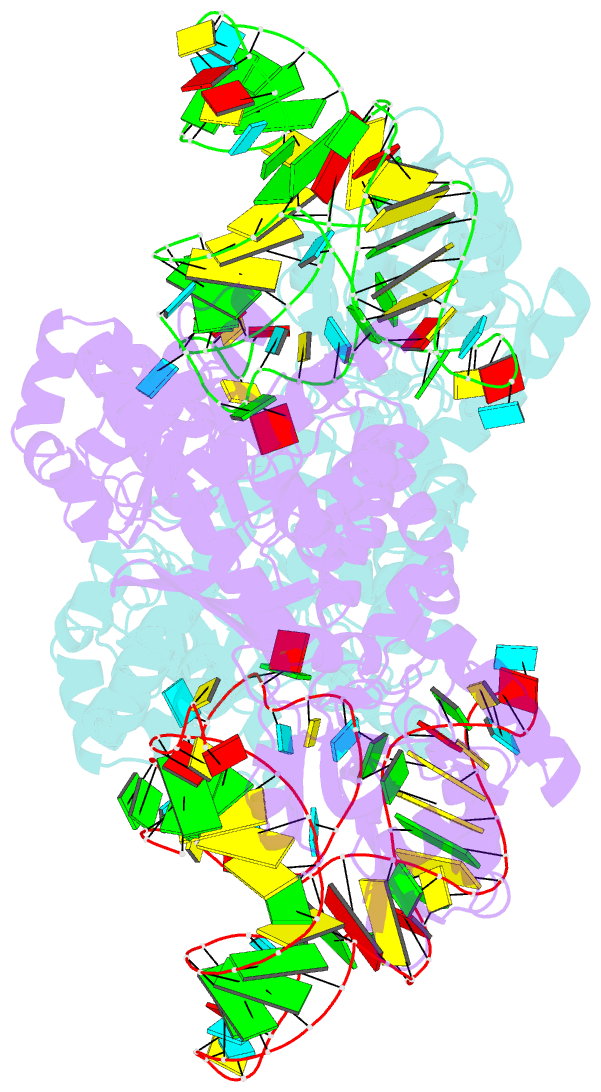
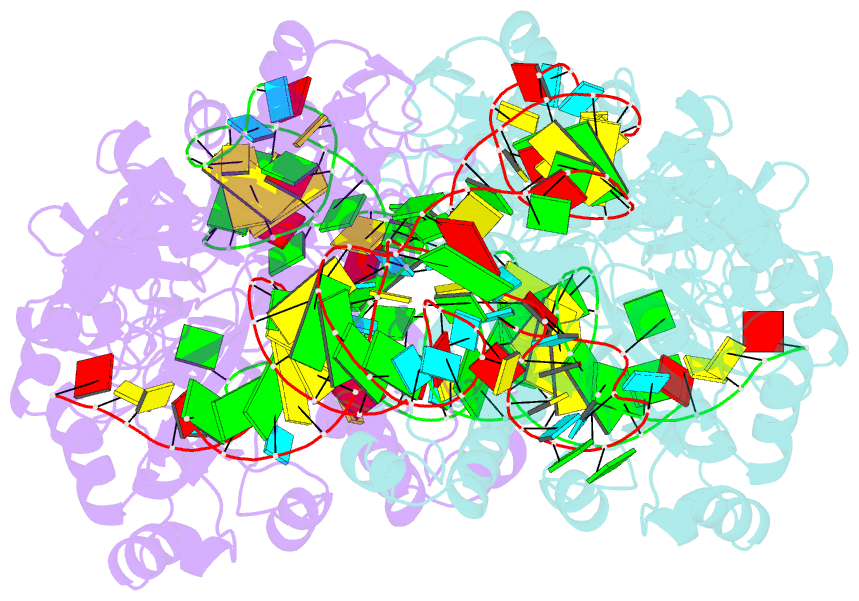
- Crystal structure of archaeosine trna-guanine transglycosylase complexed with lambda-form trna(val)
- Reference
- Ishitani R, Nureki O, Nameki N, Okada N, Nishimura S, Yokoyama S (2003): "Alternative Tertiary Structure of tRNA for Recognition by a Posttranscriptional Modification Enzyme." Cell(Cambridge,Mass.), 113, 383-394. doi: 10.1016/S0092-8674(03)00280-0.
- Abstract
- Transfer RNA (tRNA) canonically has the clover-leaf secondary structure with the acceptor, D, anticodon, and T arms, which are folded into the L-shaped tertiary structure. To strengthen the L form, posttranscriptional modifications occur on nucleotides buried within the core, but the modification enzymes are paradoxically inaccessible to them in the L form. In this study, we determined the crystal structure of tRNA bound with archaeosine tRNA-guanine transglycosylase, which modifies G15 of the D arm in the core. The bound tRNA assumes an alternative conformation ("lambda form") drastically different from the L form. All of the D-arm secondary base pairs and the canonical tertiary interactions are disrupted. Furthermore, a helical structure is reorganized, while the rest of the D arm is single stranded and protruded. Consequently, the enzyme precisely locates the exposed G15 in the active site, by counting the nucleotide number from G1 to G15 in the lambda form.