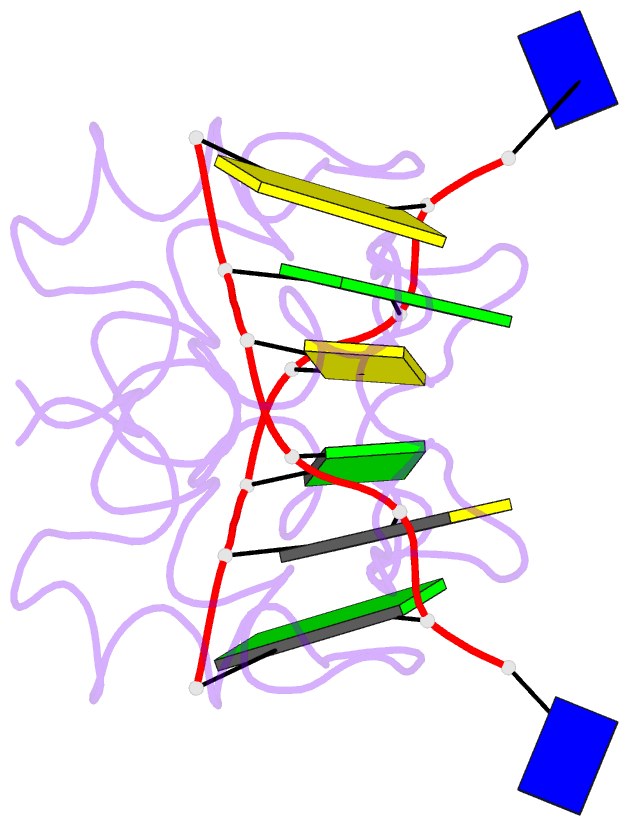
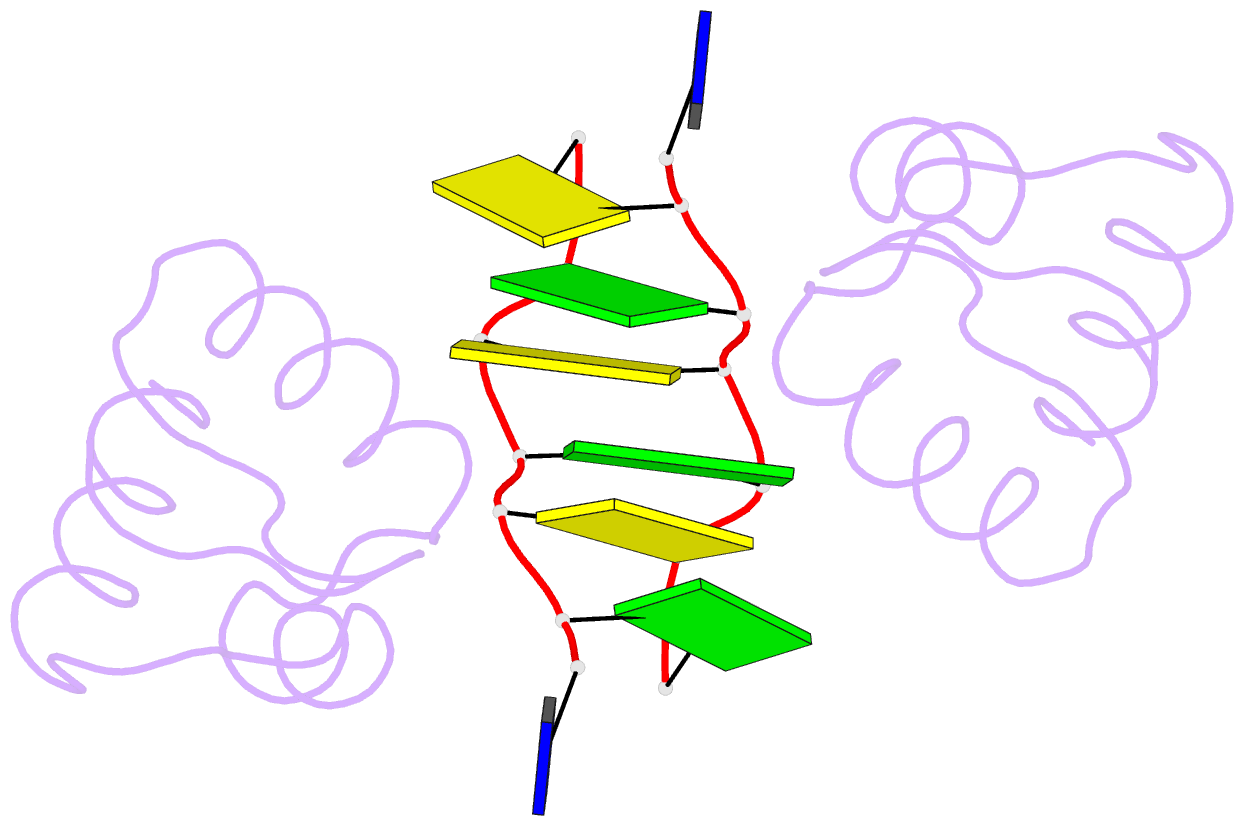
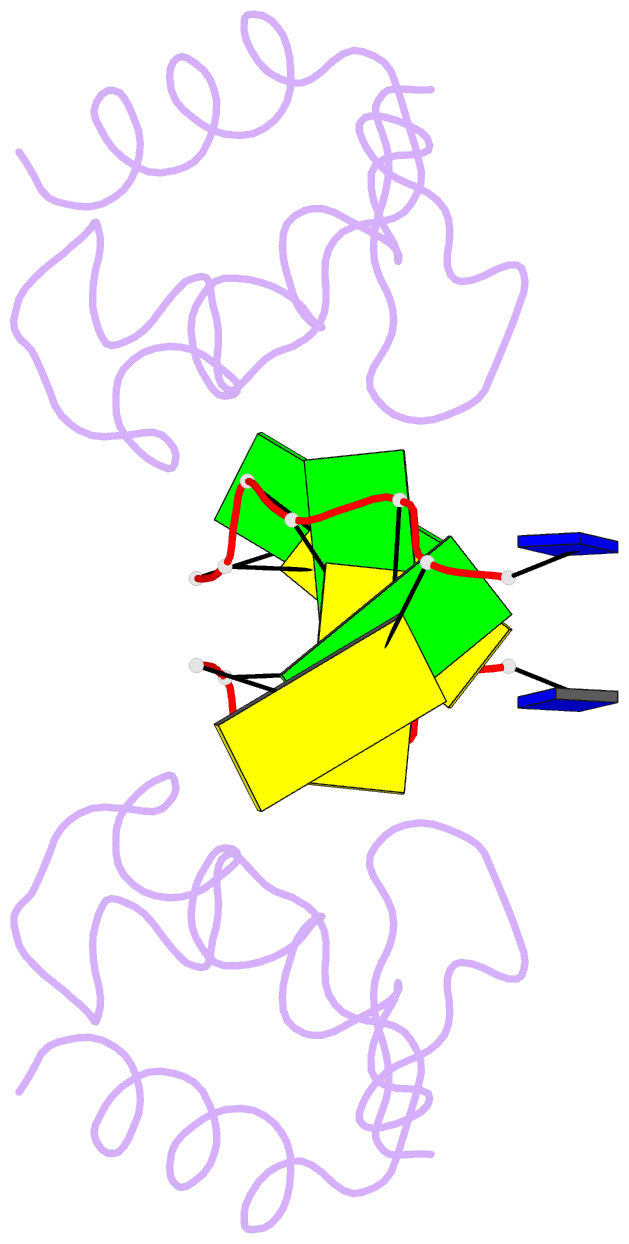
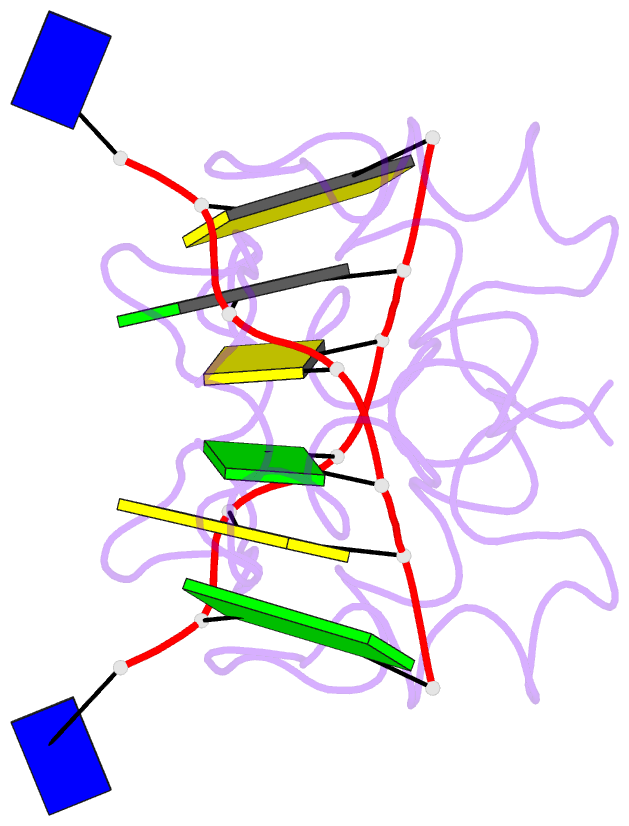
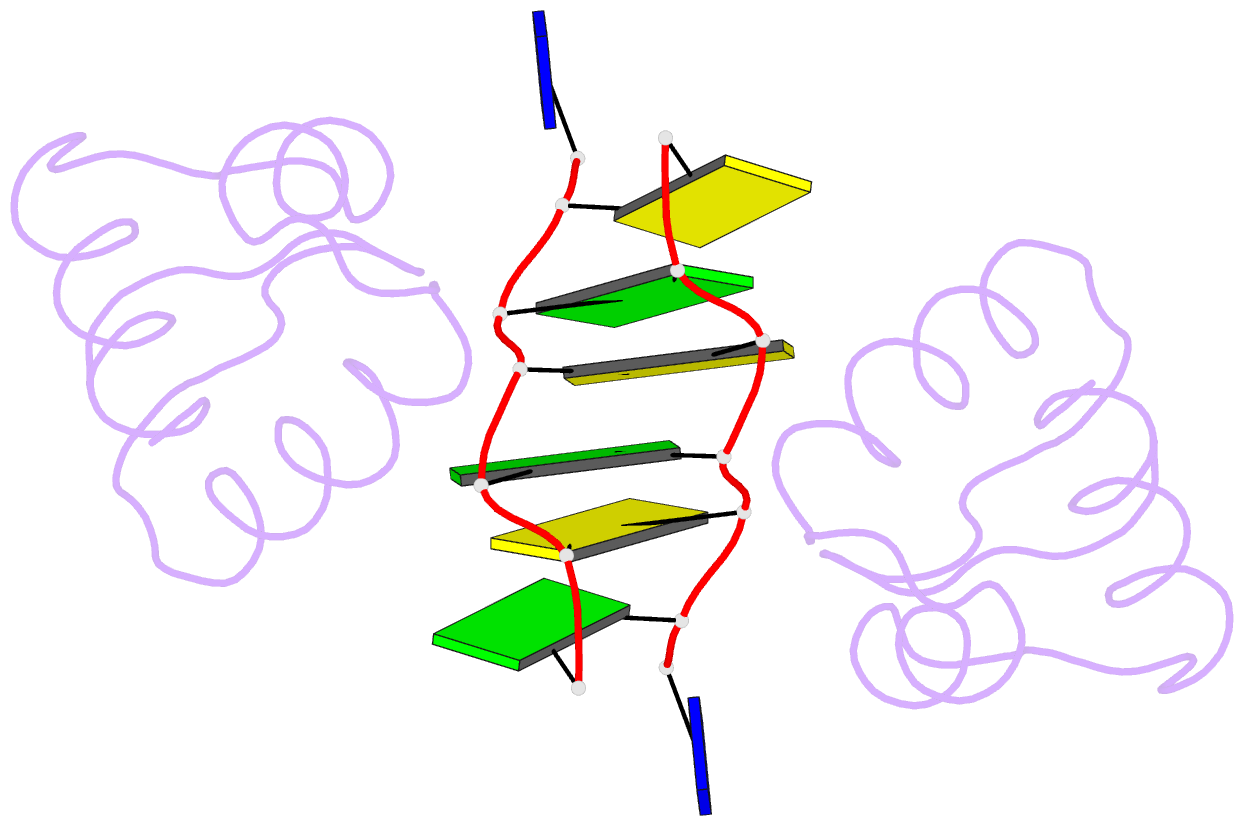
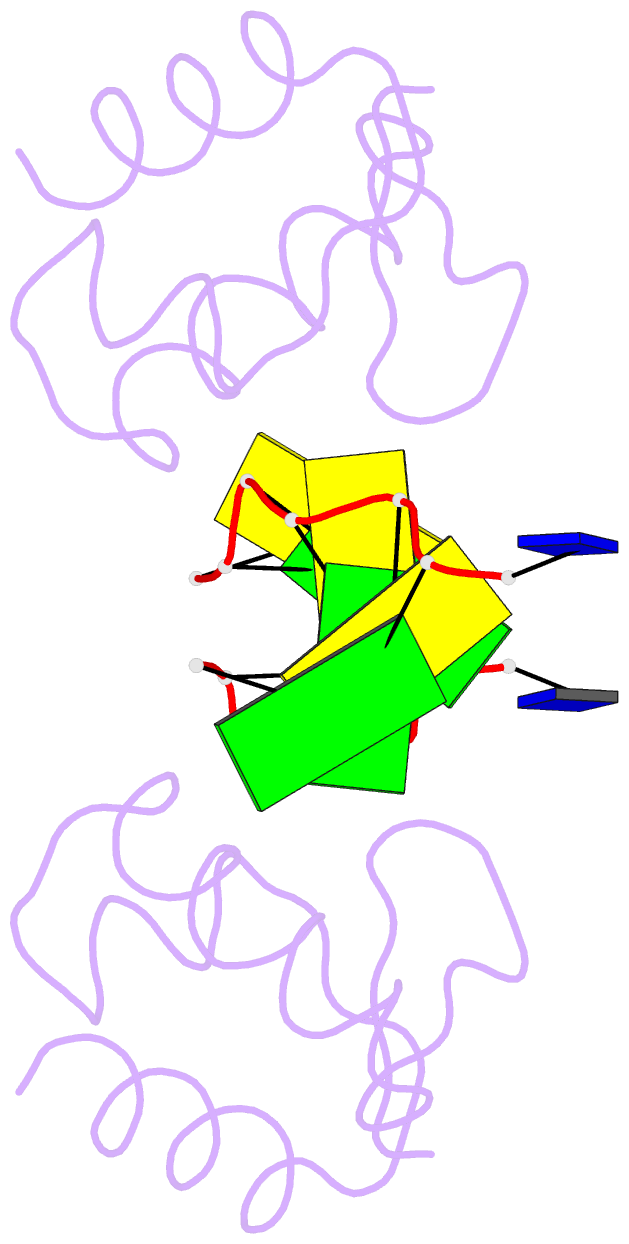
Summary information and primary citation
- PDB-id
- 1j75; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- immune system-DNA
- Method
- X-ray (1.85 Å)
- Summary
- Crystal structure of the DNA-binding domain zalpha of dlm-1 bound to z-DNA
- Reference
- Schwartz T, Behlke J, Lowenhaupt K, Heinemann U, Rich A (2001): "Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins." Nat.Struct.Biol., 8, 761-765. doi: 10.1038/nsb0901-761.
- Abstract
- The first crystal structure of a protein, the Z alpha high affinity binding domain of the RNA editing enzyme ADAR1, bound to left-handed Z-DNA was recently described. The essential set of residues determined from this structure to be critical for Z-DNA recognition was used to search the database for other proteins with the potential for Z-DNA binding. We found that the tumor-associated protein DLM-1 contains a domain with remarkable sequence similarities to Z alpha(ADAR). Here we report the crystal structure of this DLM-1 domain bound to left-handed Z-DNA at 1.85 A resolution. Comparison of Z-DNA binding by DLM-1 and ADAR1 reveals a common structure-specific recognition core within the binding domain. However, the domains differ in certain residues peripheral to the protein-DNA interface. These structures reveal a general mechanism of Z-DNA recognition, suggesting the existence of a family of winged-helix proteins sharing a common Z-DNA binding motif.