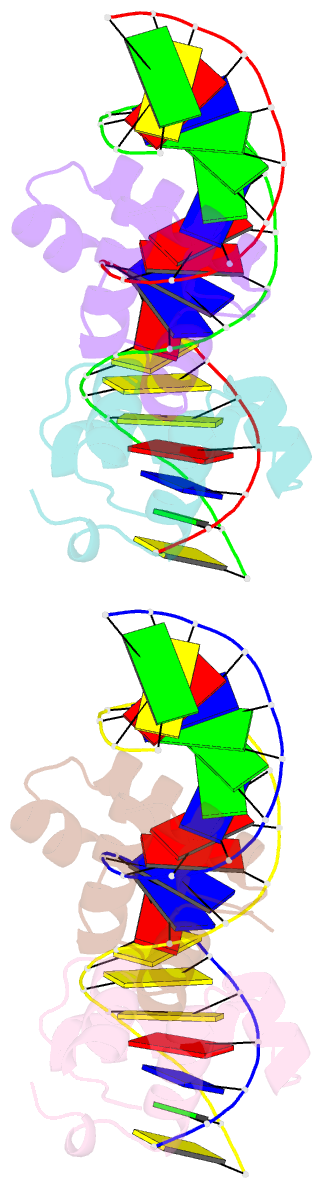
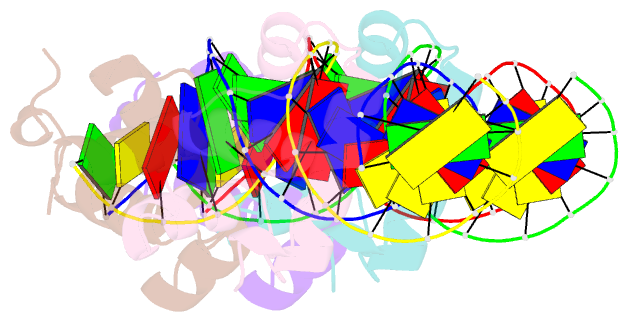
Summary information and primary citation
- PDB-id
- 1je8; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.12 Å)
- Summary
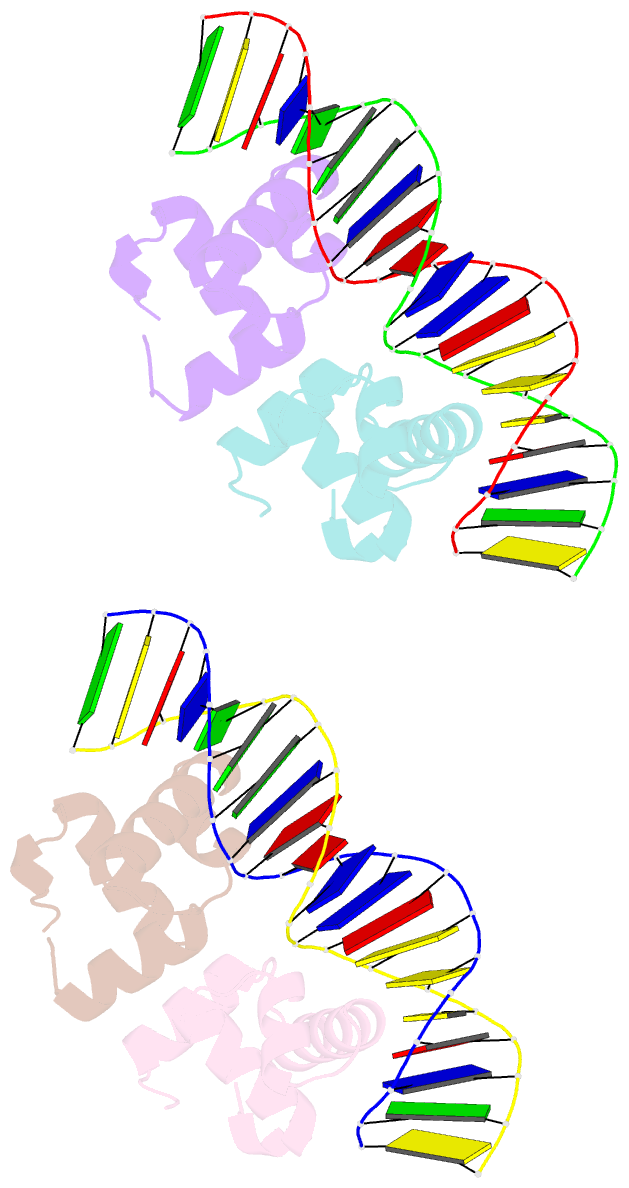
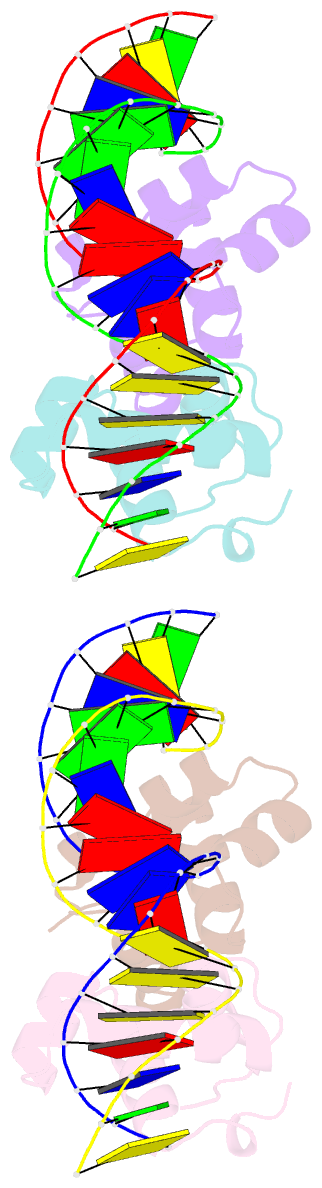
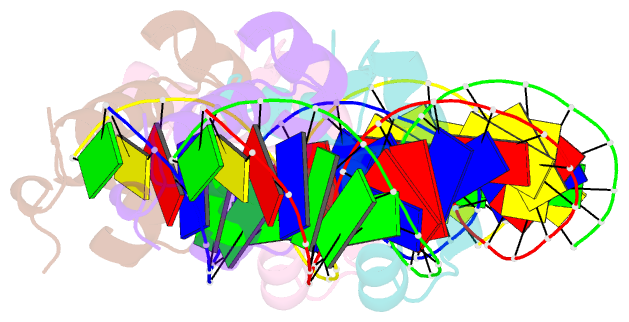
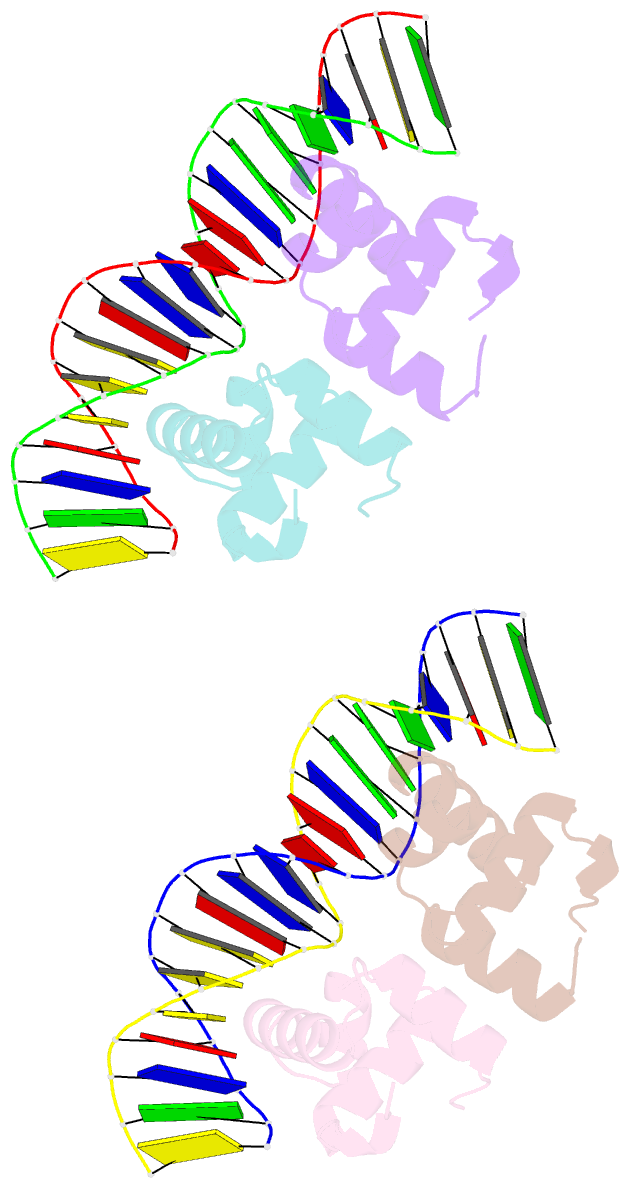
- Two-component response regulator narl-DNA complex: DNA bending found in a high affinity site
- Reference
- Maris AE, Sawaya MR, Kaczor-Grzeskowiak M, Jarvis MR, Bearson SM, Kopka ML, Schroder I, Gunsalus RP, Dickerson RE (2002): "Dimerization allows DNA target site recognition by the NarL response regulator." Nat.Struct.Biol., 9, 771-778. doi: 10.1038/nsb845.
- Abstract
- Two-component signal transduction systems are modular phosphorelay regulatory pathways common in prokaryotes. In the co-crystal structure of the Escherichia coli NarL signal output domain bound to DNA, we observe how the NarL family of two-component response regulators can bind DNA. DNA recognition is accompanied by the formation of a new dimerization interface, which could occur only in the full-length protein via a large intramolecular domain rearrangement. The DNA is recognized by the concerted effects of solvation, van der Waals forces and inherent DNA deformability, rather than determined primarily by major groove hydrogen bonding. These subtle forces permit a small DNA-binding domain to perturb the DNA helix, leading to major DNA curvature and a transition from B- to A-form DNA at the binding site, where valine on the recognition helix interacts unexpectedly with the polar major groove floor.