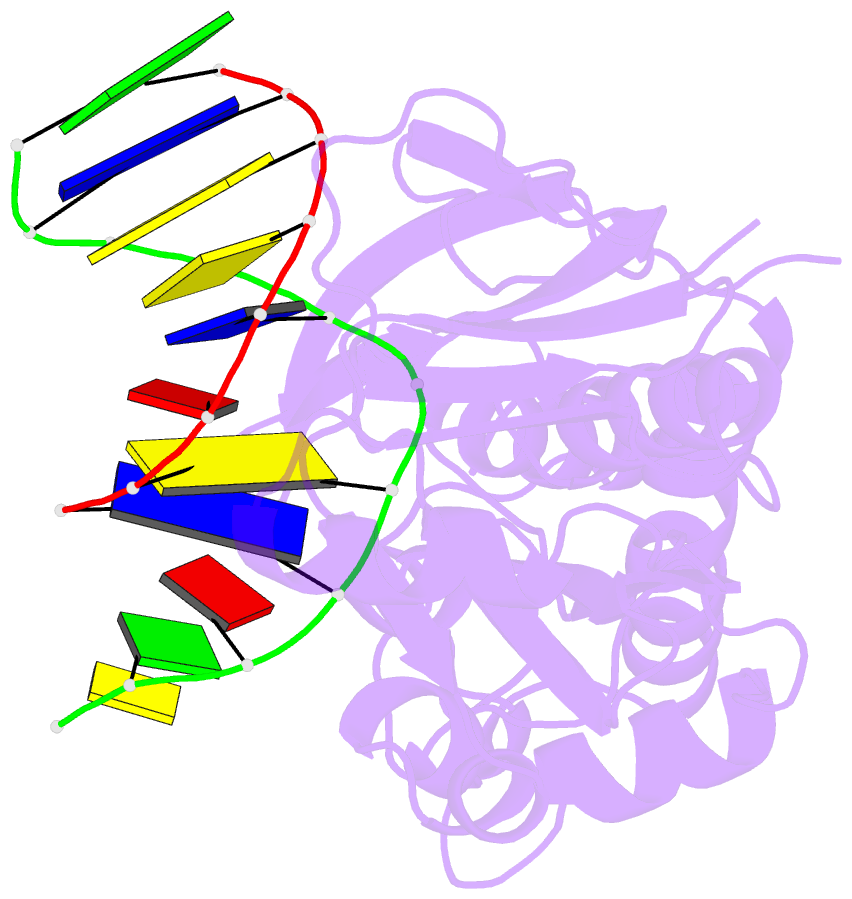
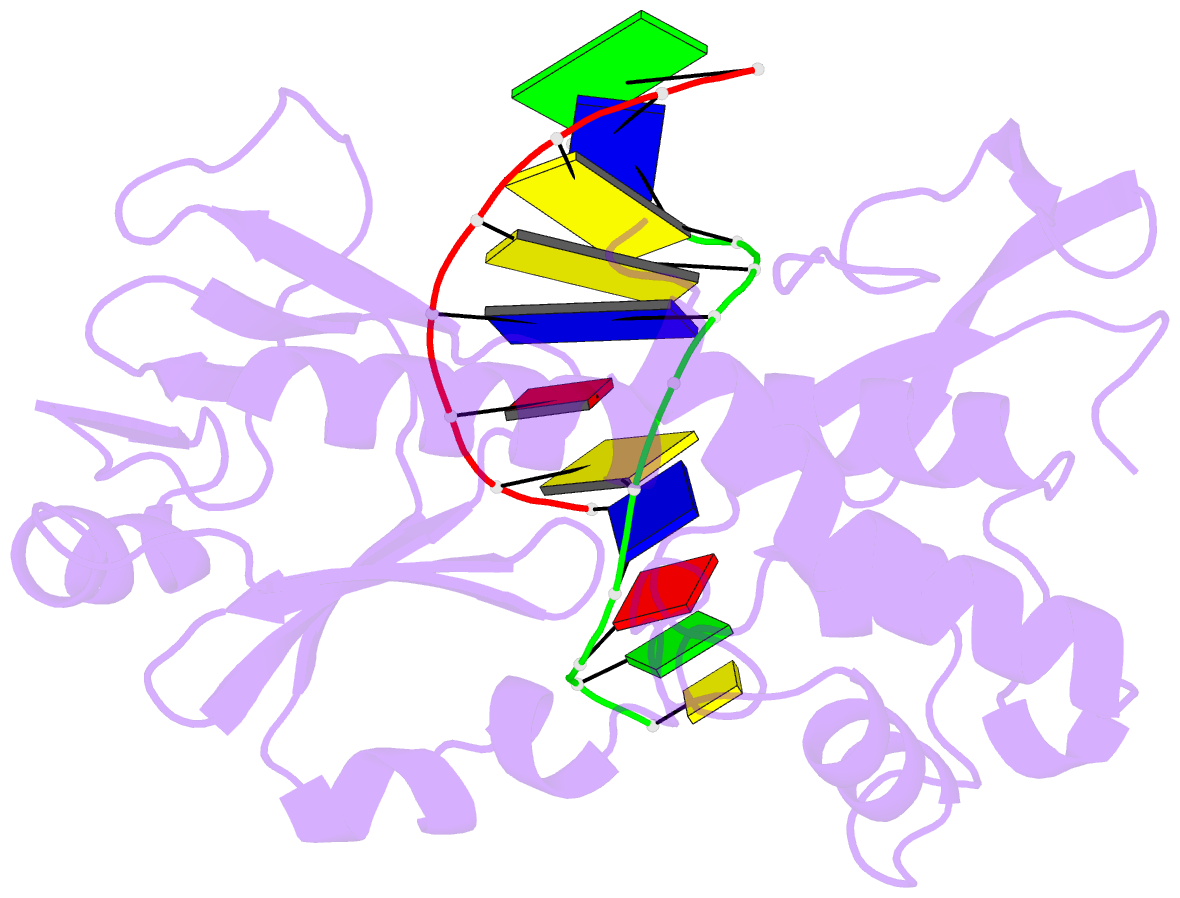
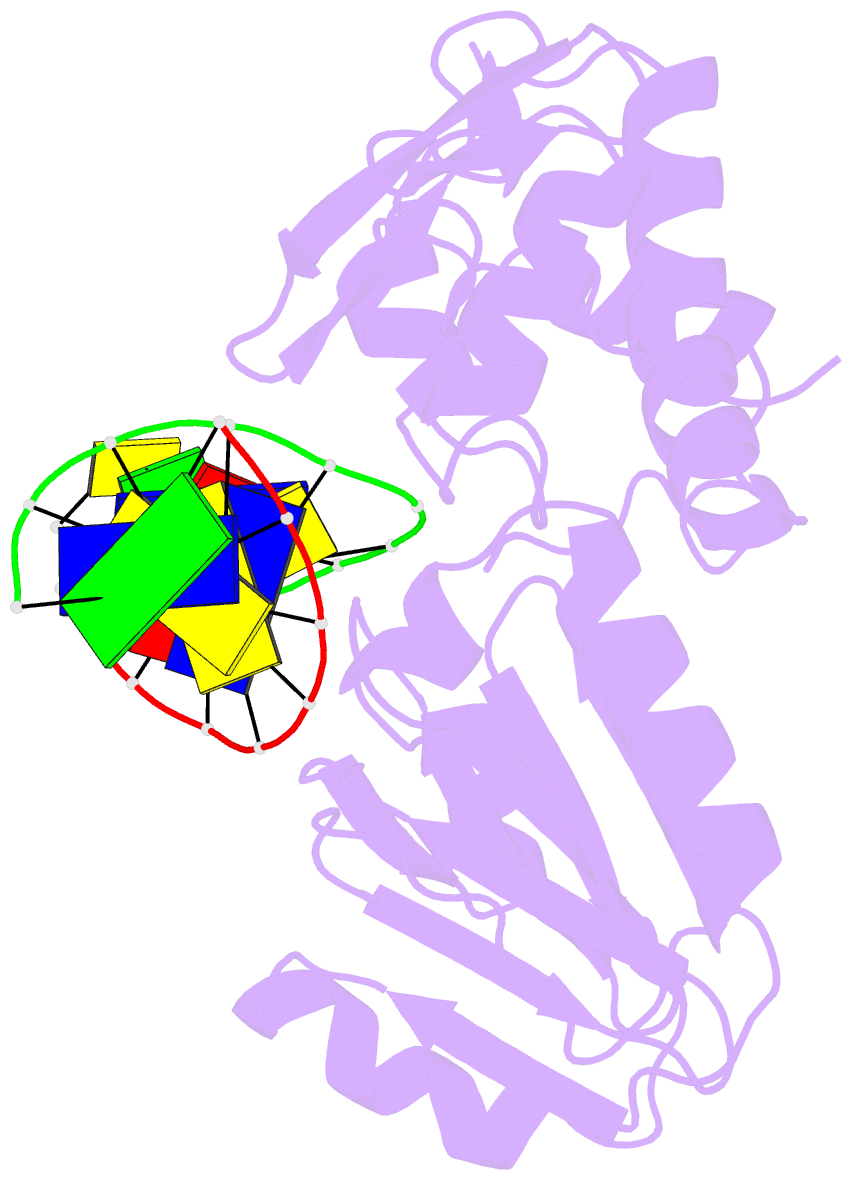
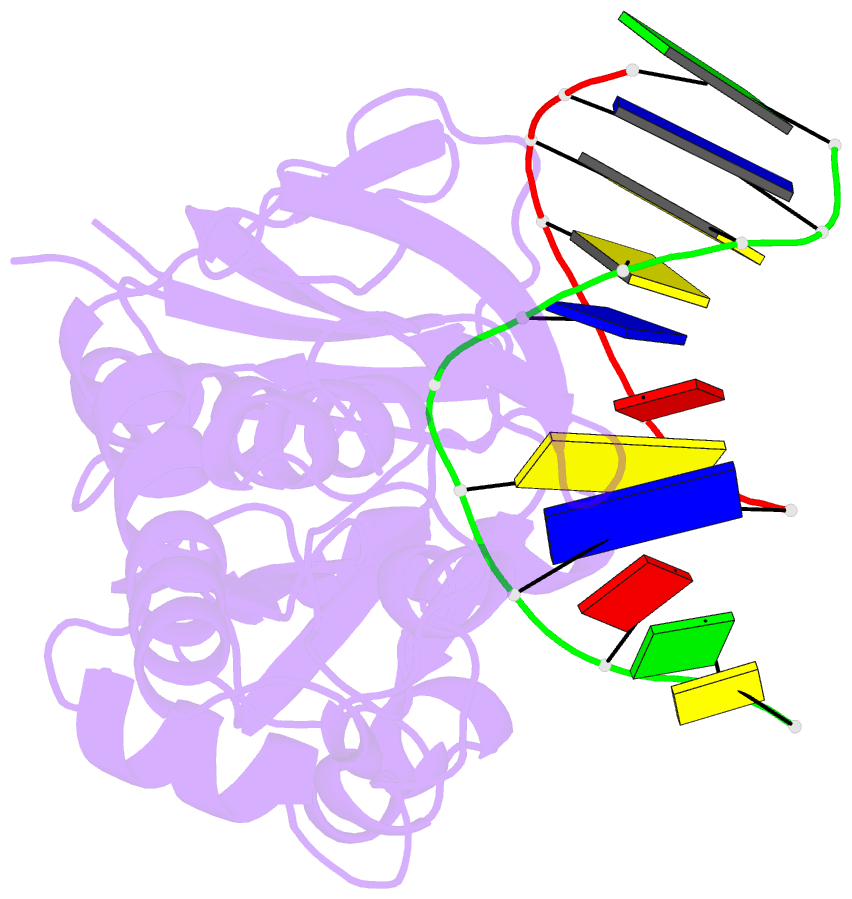
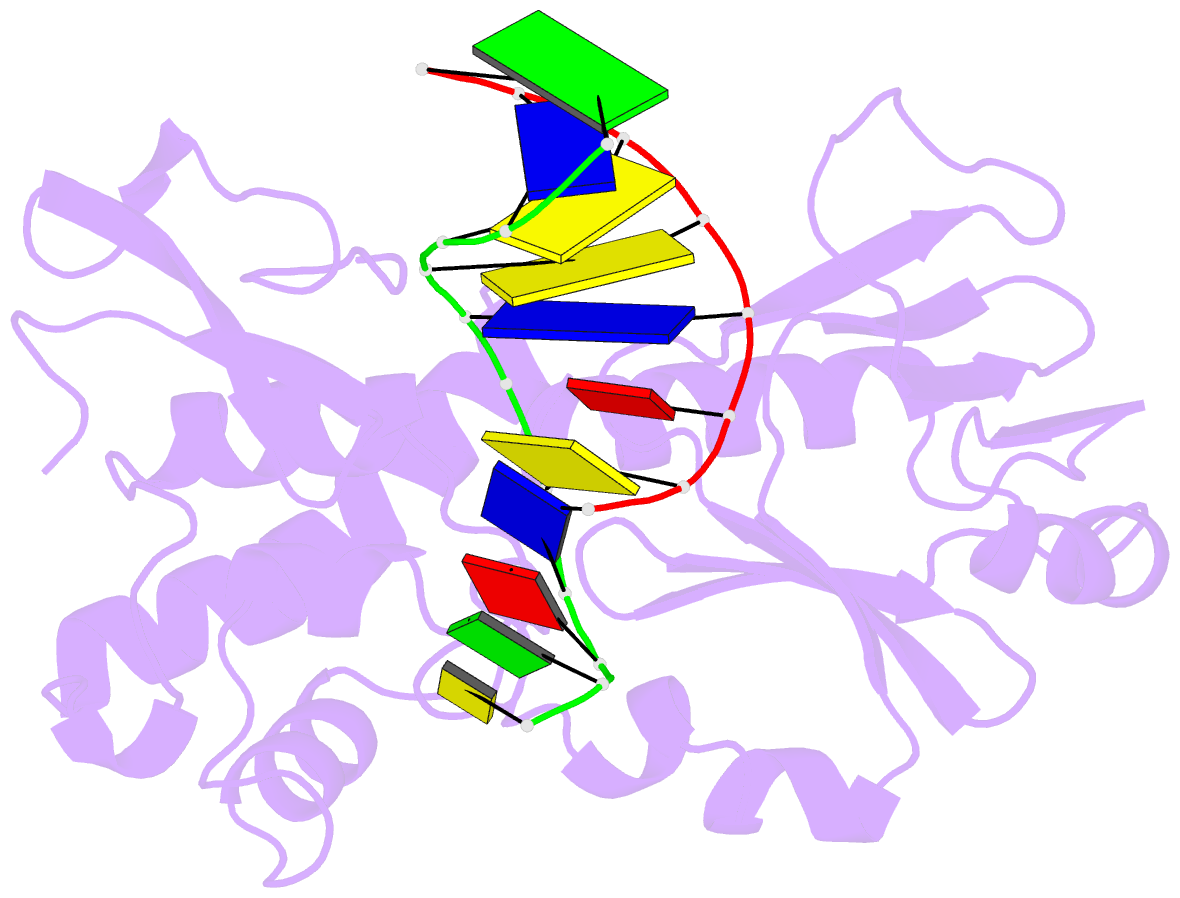
Summary information and primary citation
- PDB-id
- 1k3w; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (1.42 Å)
- Summary
- Crystal structure of a trapped reaction intermediate of the DNA repair enzyme endonuclease viii with DNA
- Reference
- Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G (2002): "Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate." EMBO J., 21, 789-800. doi: 10.1093/emboj/21.4.789.
- Abstract
- Endonuclease VIII (Nei) of Escherichia coli is a DNA repair enzyme that excises oxidized pyrimidines from DNA. Nei shares with formamidopyrimidine-DNA glycosylase (Fpg) sequence homology and a similar mechanism of action: the latter involves removal of the damaged base followed by two sequential beta-elimination steps. However, Nei differs significantly from Fpg in substrate specificity. We determined the structure of Nei covalently crosslinked to a 13mer oligodeoxynucleotide duplex at 1.25 A resolution. The crosslink is derived from a Schiff base intermediate that precedes beta-elimination and is stabilized by reduction with NaBH(4). Nei consists of two domains connected by a hinge region, creating a DNA binding cleft between domains. DNA in the complex is sharply kinked, the deoxyribitol moiety is bound covalently to Pro1 and everted from the duplex into the active site. Amino acids involved in substrate binding and catalysis are identified. Molecular modeling and analysis of amino acid conservation suggest a site for recognition of the damaged base. Based on structural features of the complex and site-directed mutagenesis studies, we propose a catalytic mechanism for Nei.