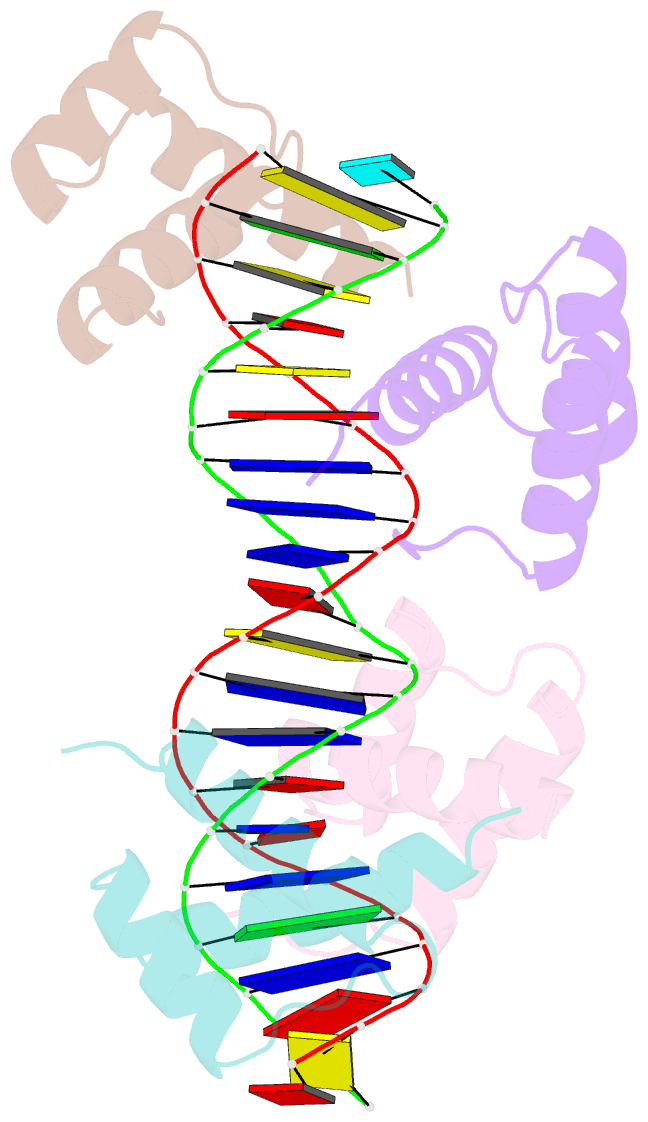
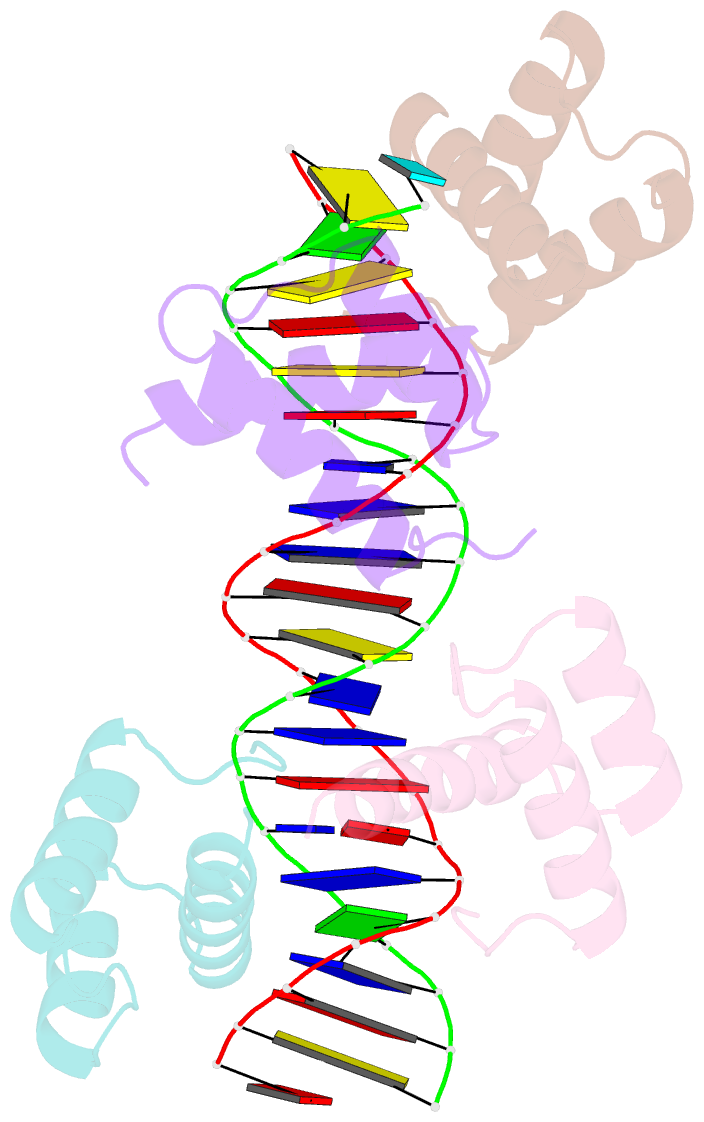
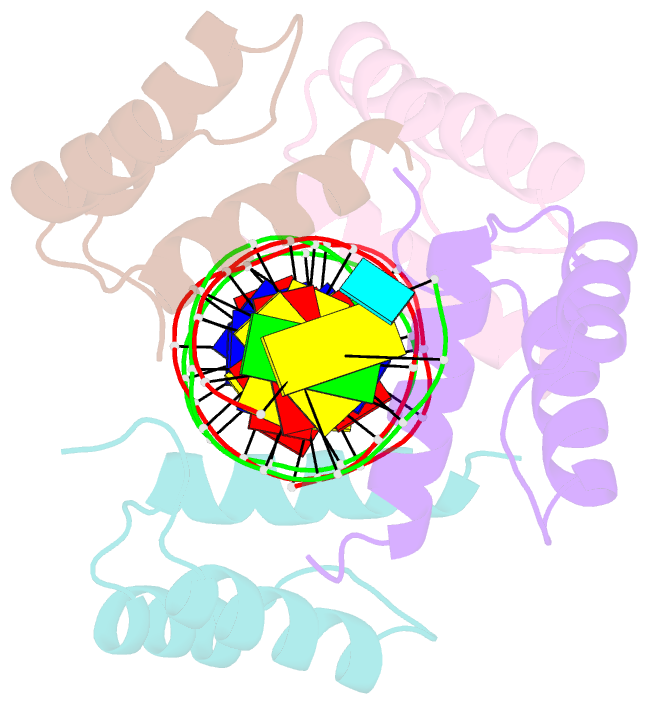
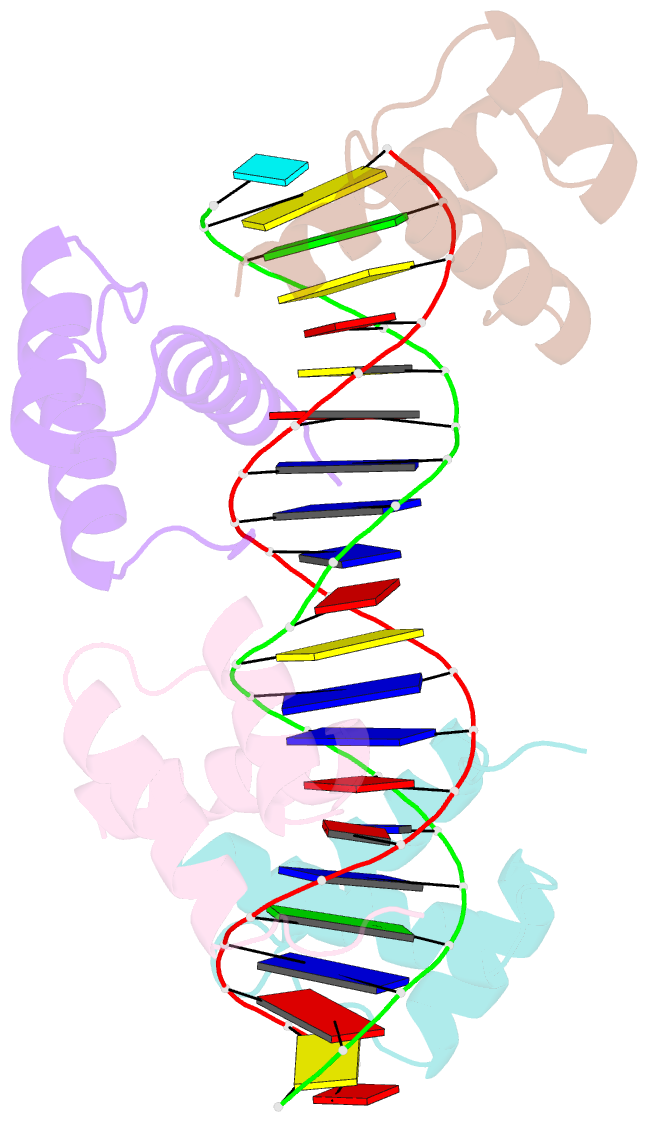
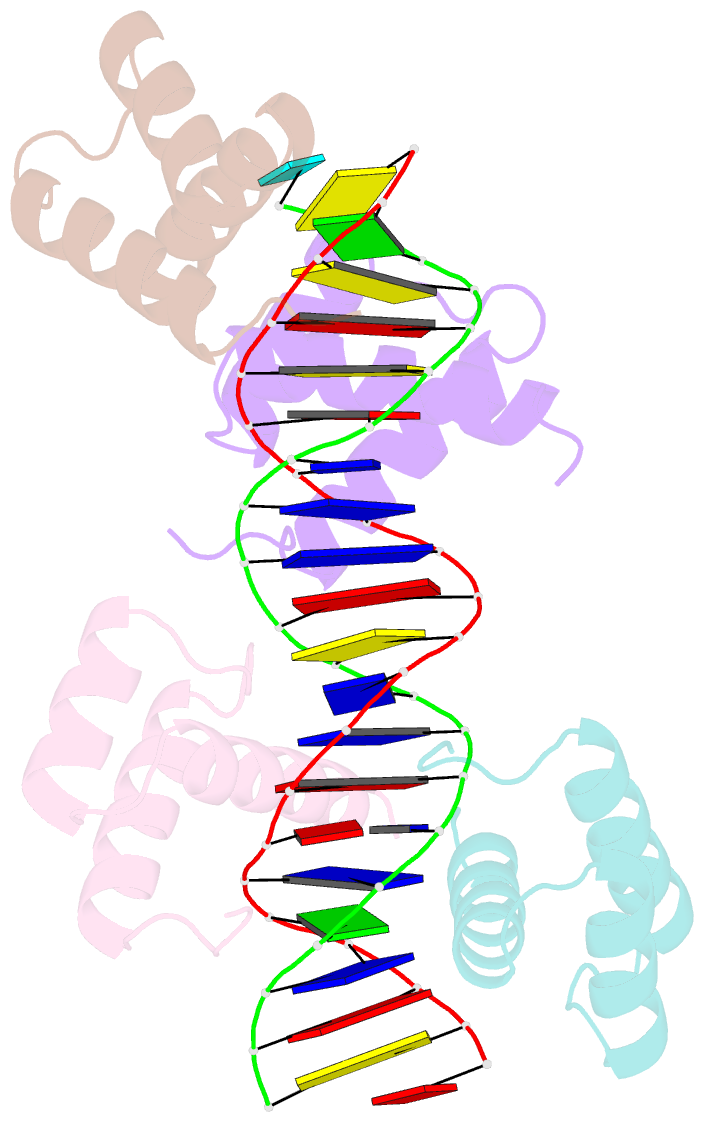
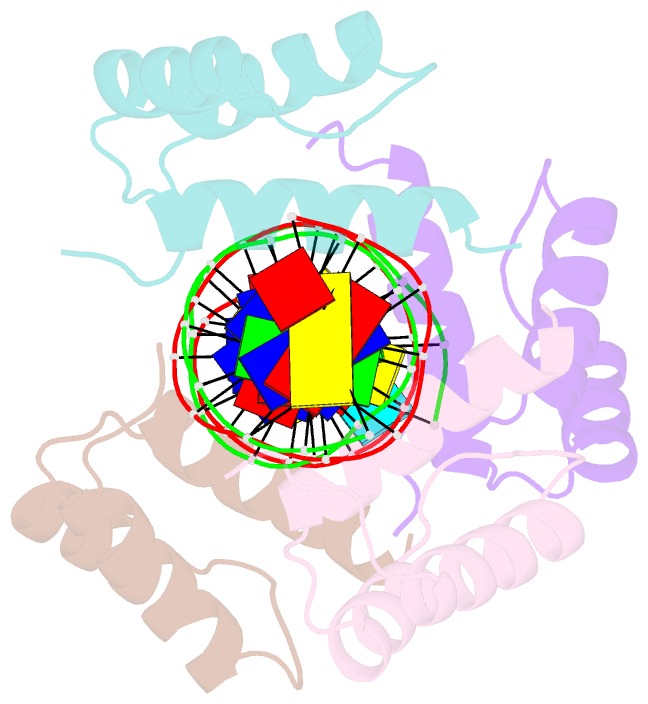
Summary information and primary citation
- PDB-id
- 1k61; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.1 Å)
- Summary
- Matalpha2 homeodomain bound to DNA
- Reference
- Aishima J, Gitti RK, Noah JE, Gan HH, Schlick T, Wolberger C (2002): "A Hoogsteen base pair embedded in undistorted B-DNA." NUCLEIC ACIDS RES., 30, 5244-5252. doi: 10.1093/nar/gkf661.
- Abstract
- Hoogsteen base pairs within duplex DNA typically are only observed in regions containing significant distortion or near sites of drug intercalation. We report here the observation of a Hoogsteen base pair embedded within undistorted, unmodified B-DNA. The Hoogsteen base pair, consisting of a syn adenine base paired with an anti thymine base, is found in the 2.1 A resolution structure of the MATalpha2 homeodomain bound to DNA in a region where a specifically and a non-specifically bound homeodomain contact overlapping sites. NMR studies of the free DNA show no evidence of Hoogsteen base pair formation, suggesting that protein binding favors the transition from a Watson-Crick to a Hoogsteen base pair. Molecular dynamics simulations of the homeodomain-DNA complex support a role for the non-specifically bound protein in favoring Hoogsteen base pair formation. The presence of a Hoogsteen base pair in the crystal structure of a protein-DNA complex raises the possibility that Hoogsteen base pairs could occur within duplex DNA and play a hitherto unrecognized role in transcription, replication and other cellular processes.