Summary information and primary citation
- PDB-id
- 1k8w; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- lyase-RNA
- Method
- X-ray (1.85 Å)
- Summary
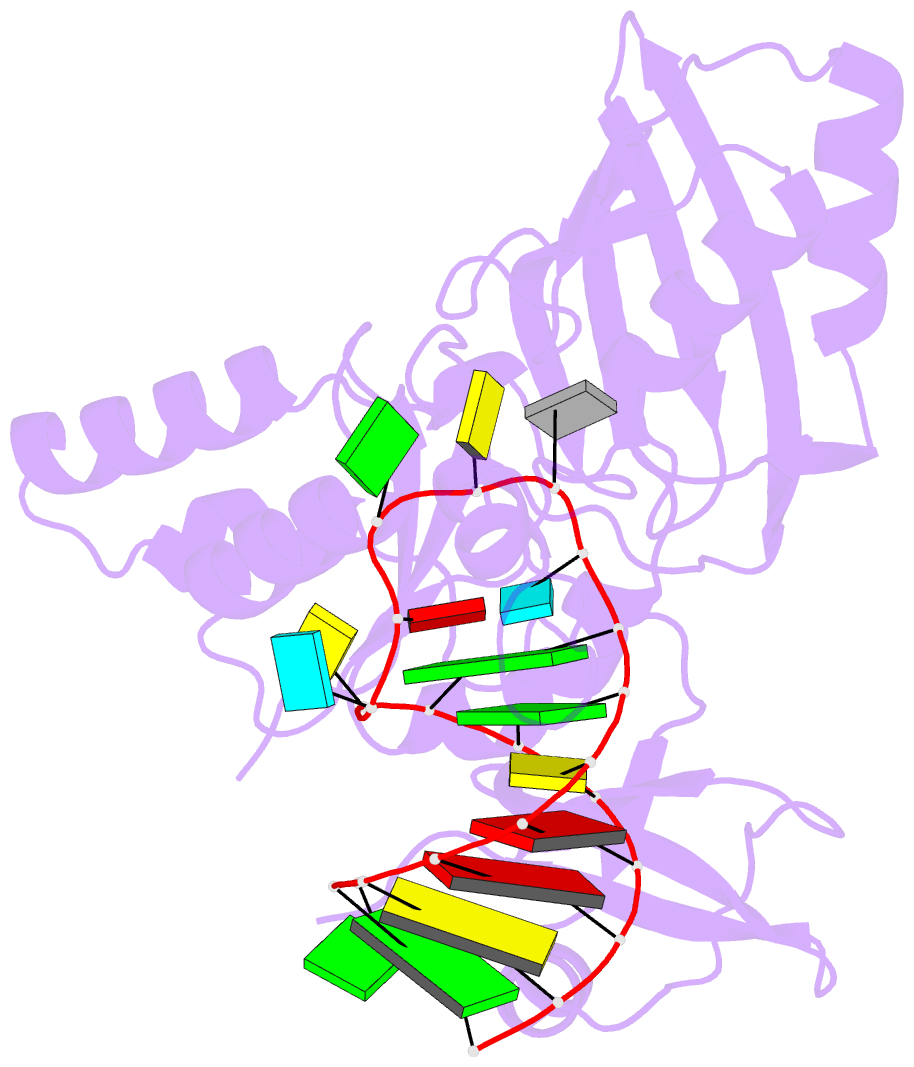
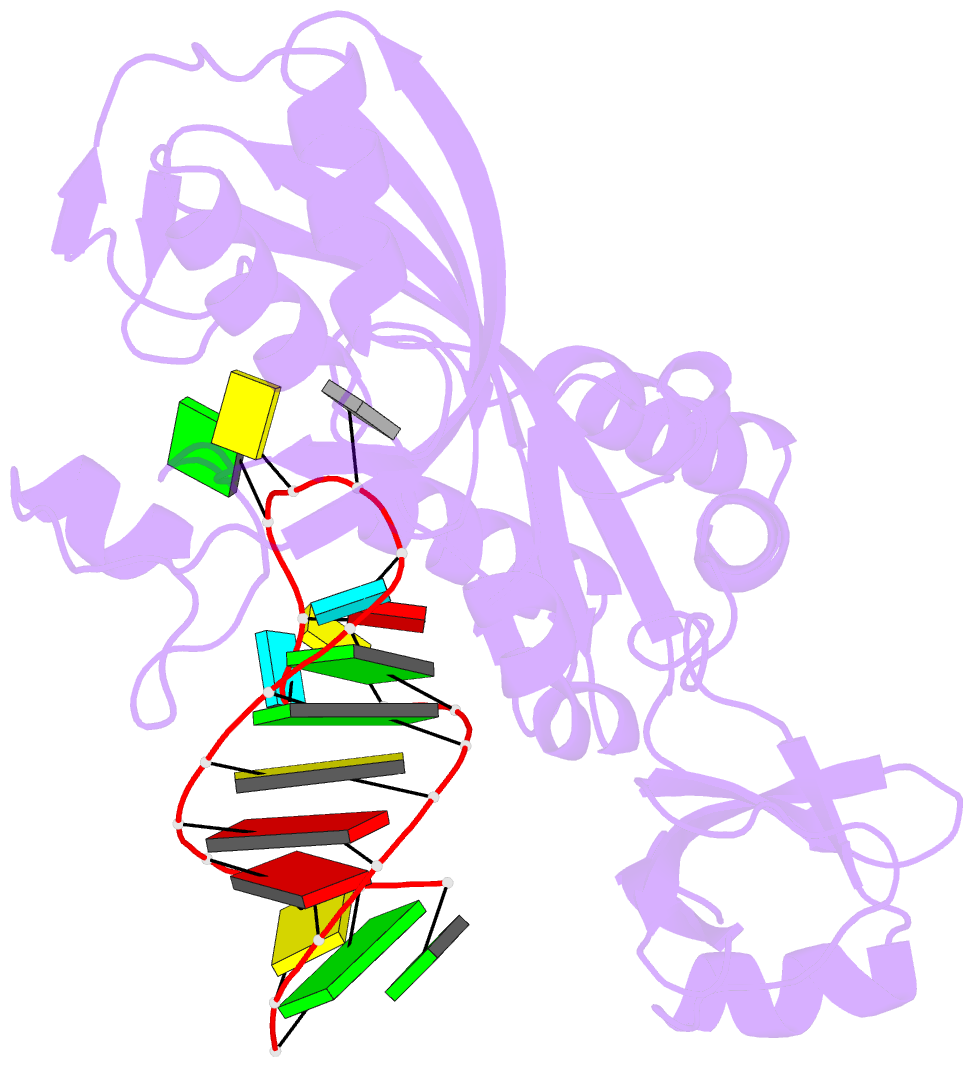
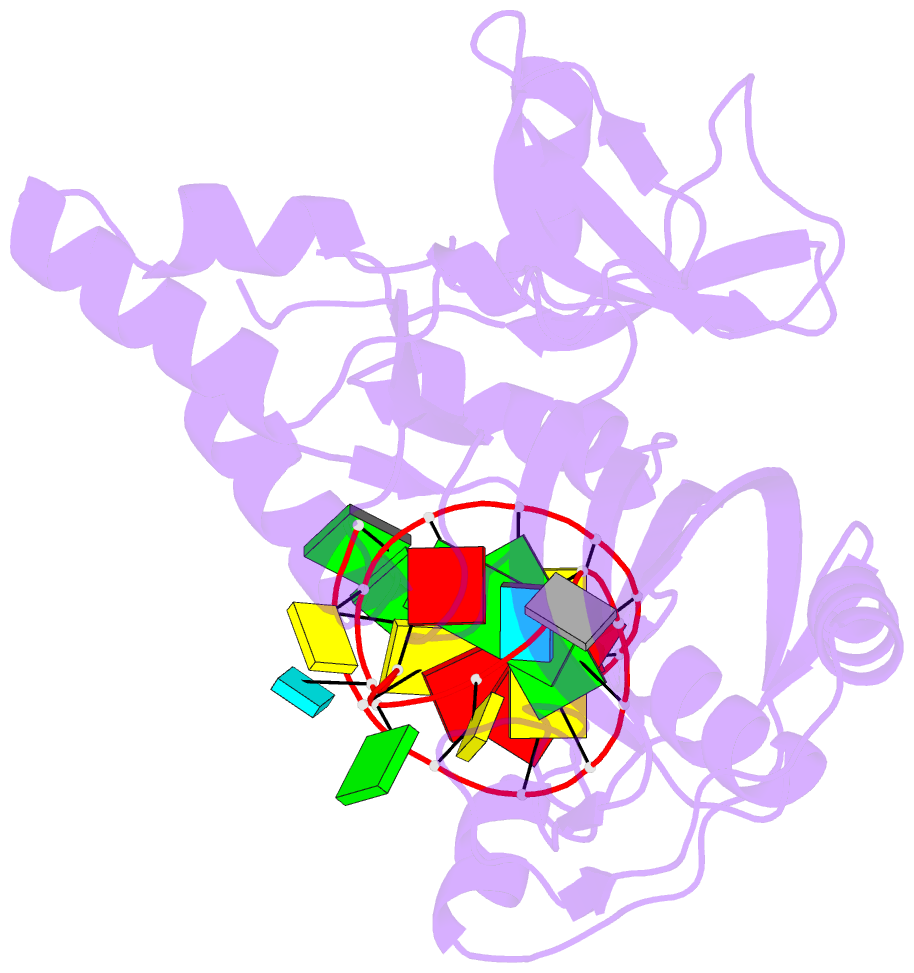
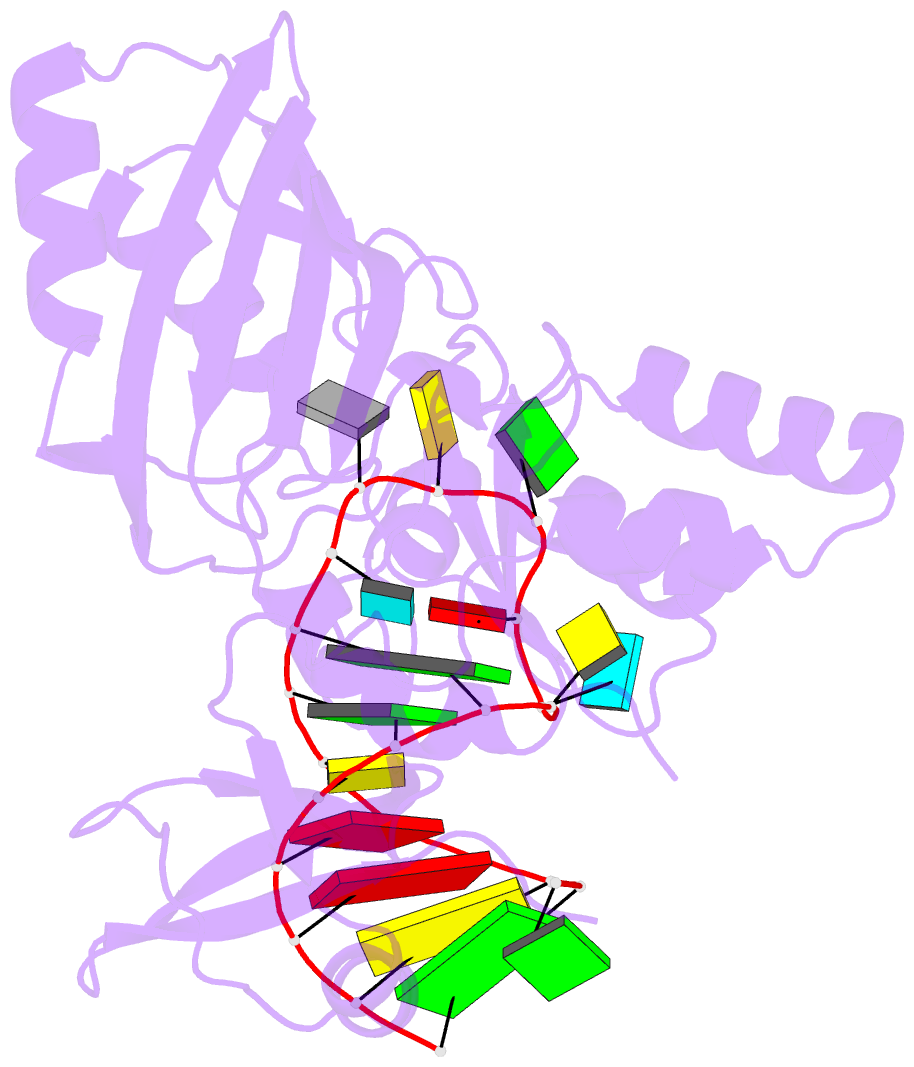
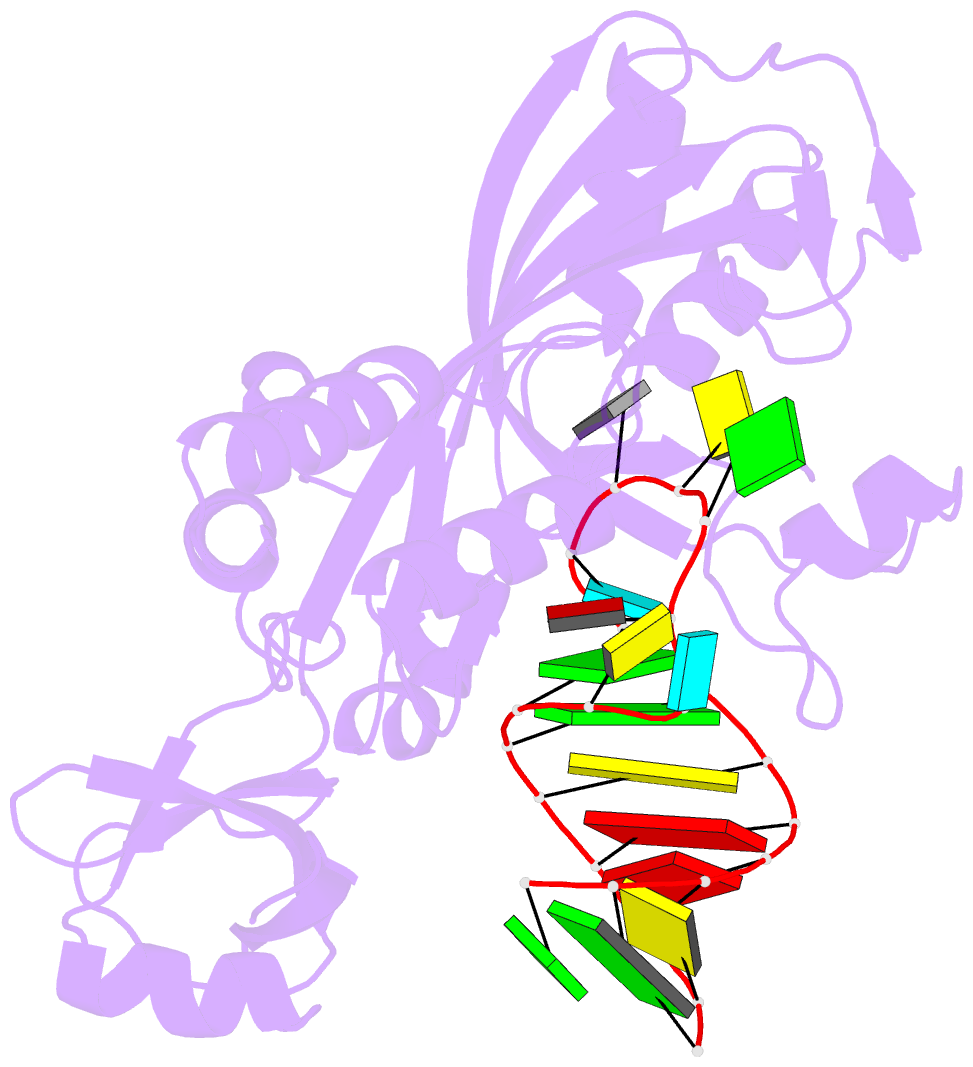
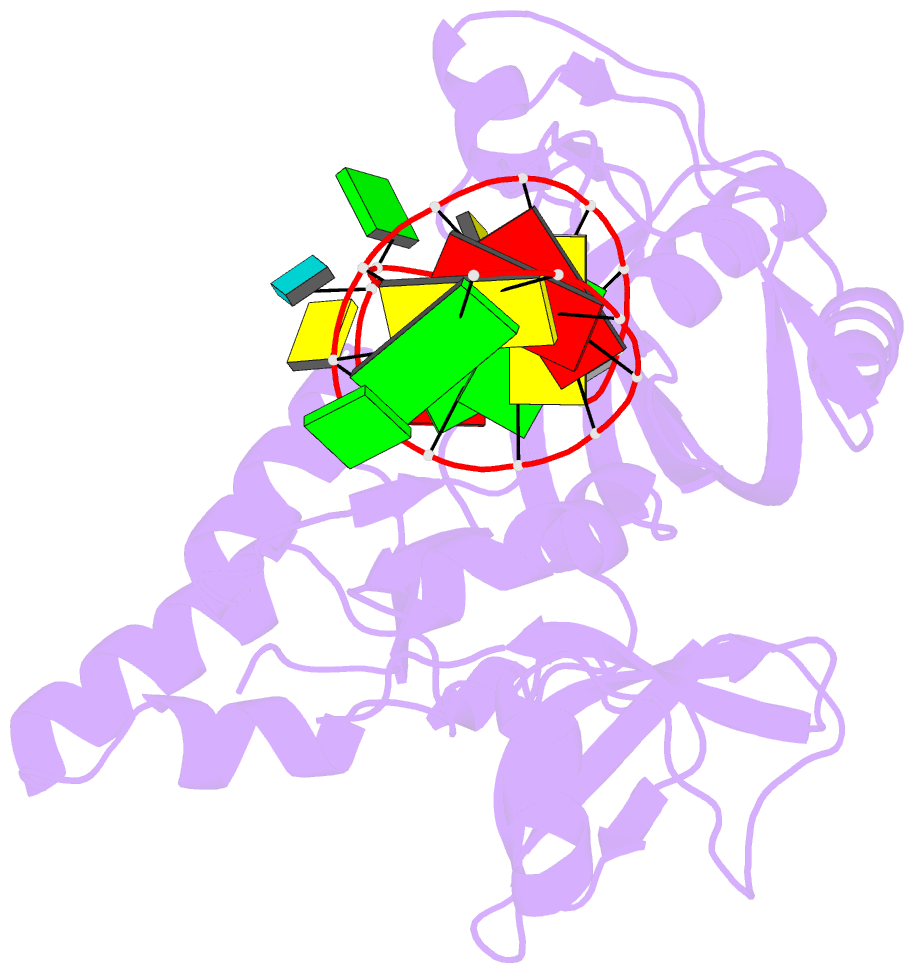
- Crystal structure of the e. coli pseudouridine synthase trub bound to a t stem-loop RNA
- Reference
- Hoang C, Ferre-D'Amare AR (2001): "Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme." Cell(Cambridge,Mass.), 107, 929-939. doi: 10.1016/S0092-8674(01)00618-3.
- Abstract
- Pseudouridine (Psi) synthases catalyze the isomerization of specific uridines in cellular RNAs to pseudouridines and may function as RNA chaperones. TruB is responsible for the Psi residue present in the T loops of virtually all tRNAs. The close homolog Cbf5/dyskerin is the catalytic subunit of box H/ACA snoRNPs. These carry out the pseudouridylation of eukaryotic rRNA and snRNAs. The 1.85 A resolution structure of TruB bound to RNA reveals that this enzyme recognizes the preformed three-dimensional structure of the T loop, primarily through shape complementarity. It accesses its substrate uridyl residue by flipping out the nucleotide and disrupts the tertiary structure of tRNA. Structural comparisons with TruB demonstrate that all Psi synthases are descended from a common molecular ancestor.