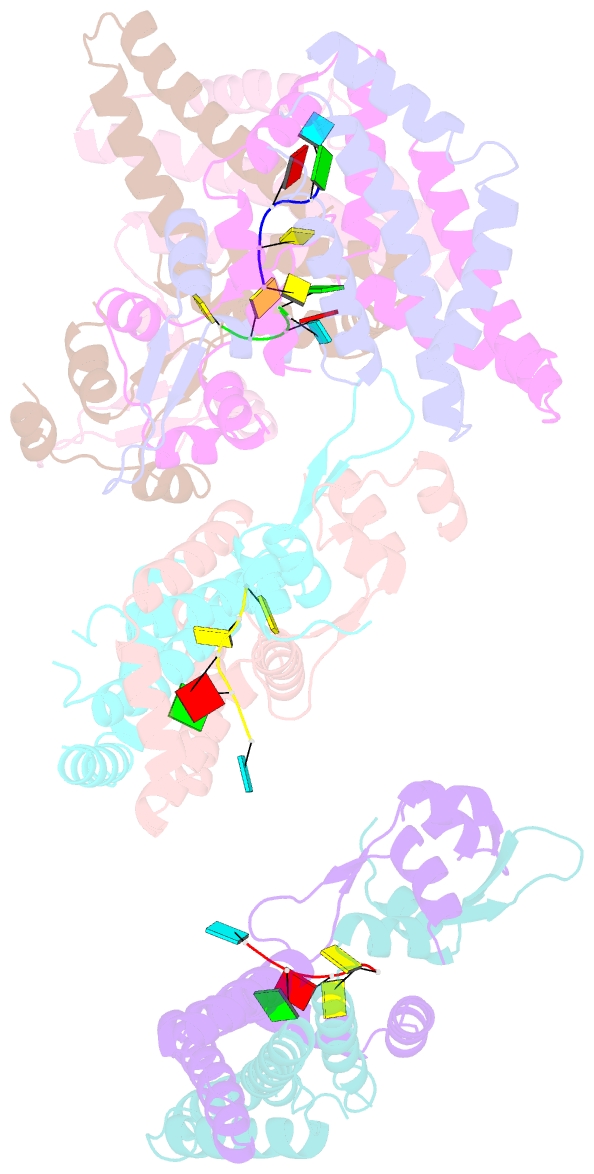
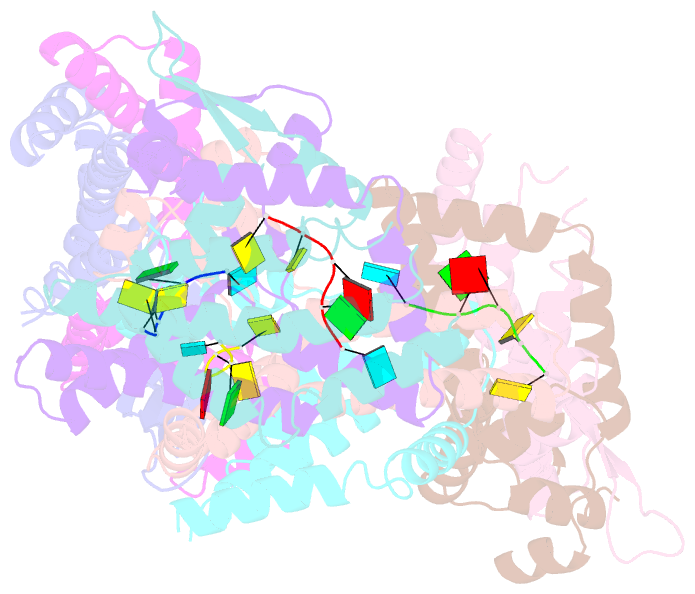
Summary information and primary citation
- PDB-id
- 1knz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- X-ray (2.45 Å)
- Summary
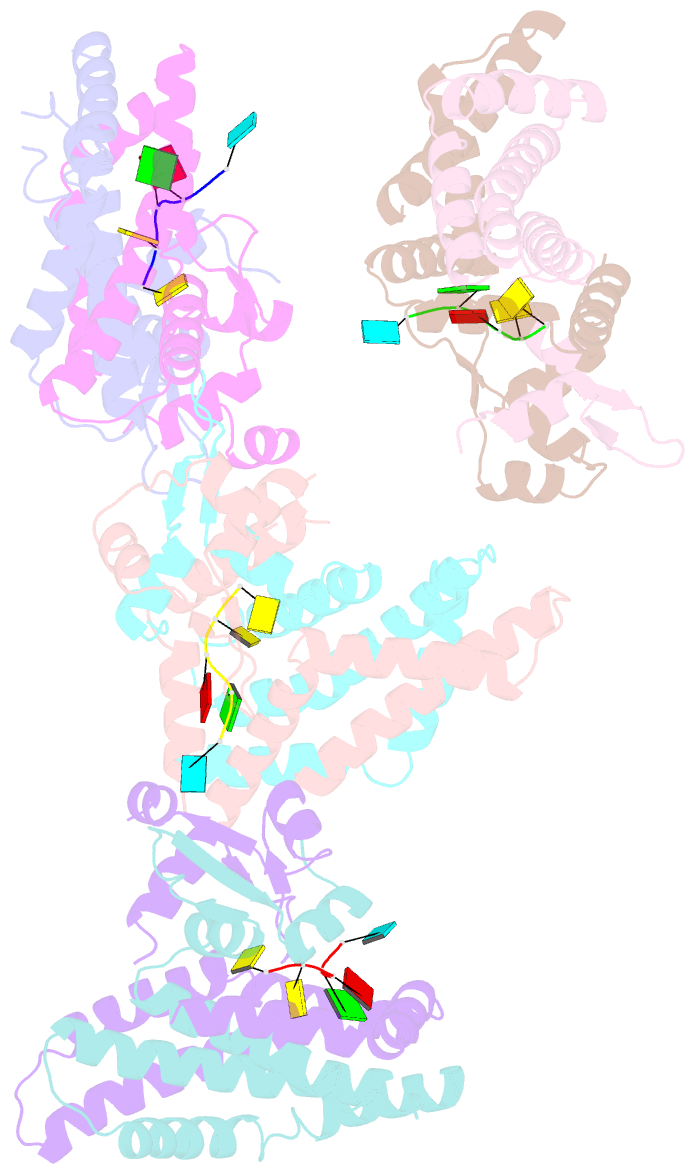
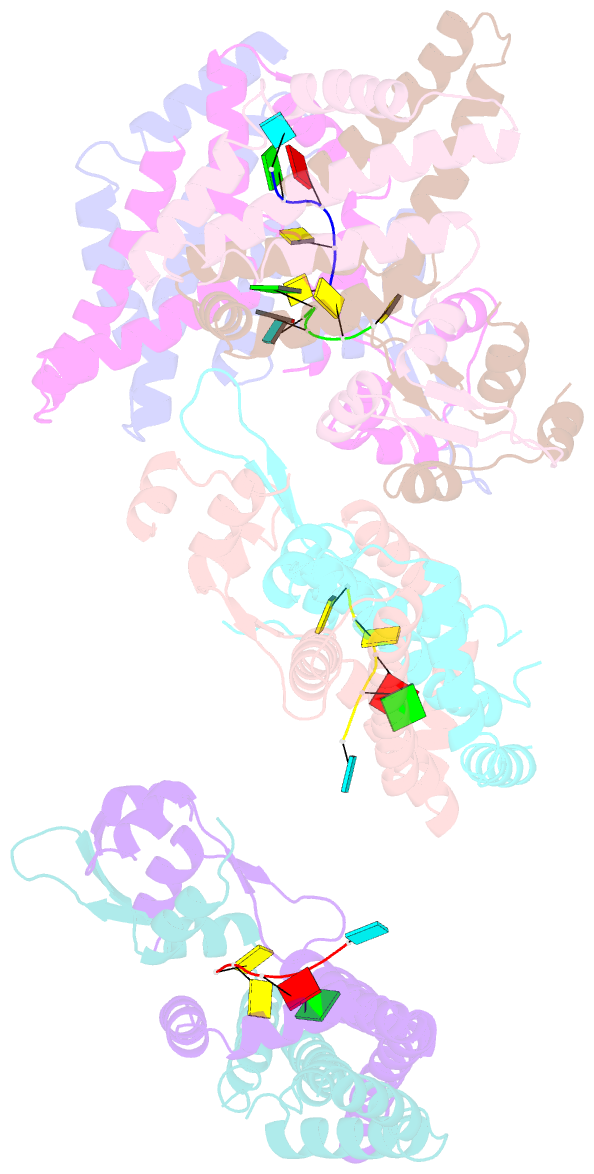
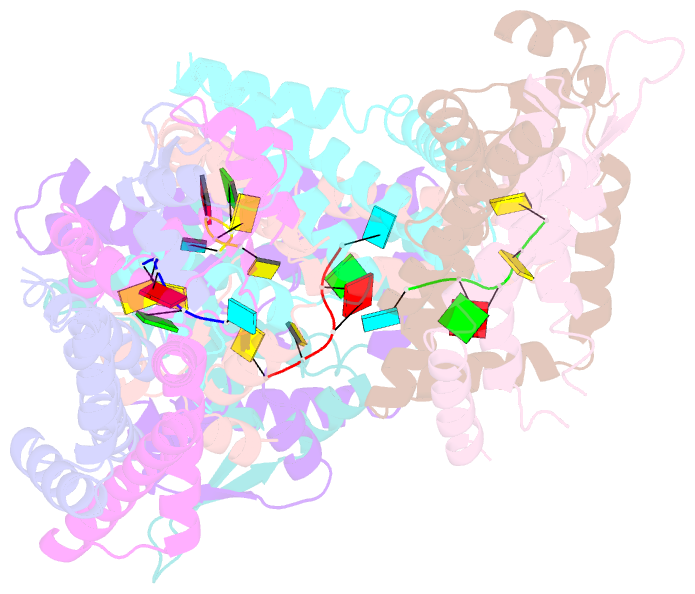
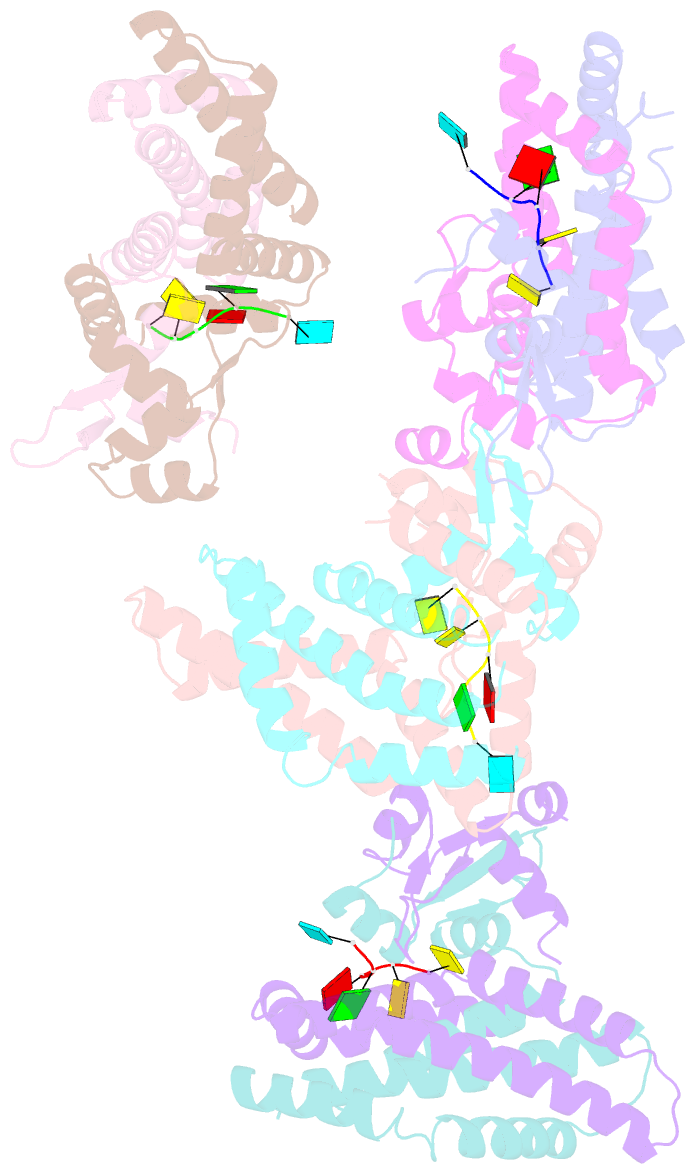
- Recognition of the rotavirus mrna 3' consensus by an asymmetric nsp3 homodimer
- Reference
- Deo RC, Groft CM, Rajashankar KR, Burley SK (2002): "Recognition of the rotavirus mRNA 3' consensus by an asymmetric NSP3 homodimer." Cell(Cambridge,Mass.), 108, 71-81. doi: 10.1016/S0092-8674(01)00632-8.
- Abstract
- Rotaviruses, the cause of life-threatening diarrhea in humans and cattle, utilize a functional homolog of poly(A) binding protein (PABP) known as nonstructural protein 3 (NSP3) for translation of viral mRNAs. NSP3 binds to viral mRNA 3' consensus sequences and circularizes the mRNA via interactions with eIF4G. The X-ray structure of the NSP3 RNA binding domain bound to a rotaviral mRNA 3' end has been determined. NSP3 is a novel, heart-shaped homodimer with a medial RNA binding cleft. The homodimer is asymmetric, and contains two similar N-terminal segments plus two structurally different C-terminal segments that intertwine to create a tunnel enveloping the mRNA 3' end. Biophysical studies demonstrate high affinity binding leading to increased thermal stability and slow dissociation kinetics, consistent with NSP3 function.