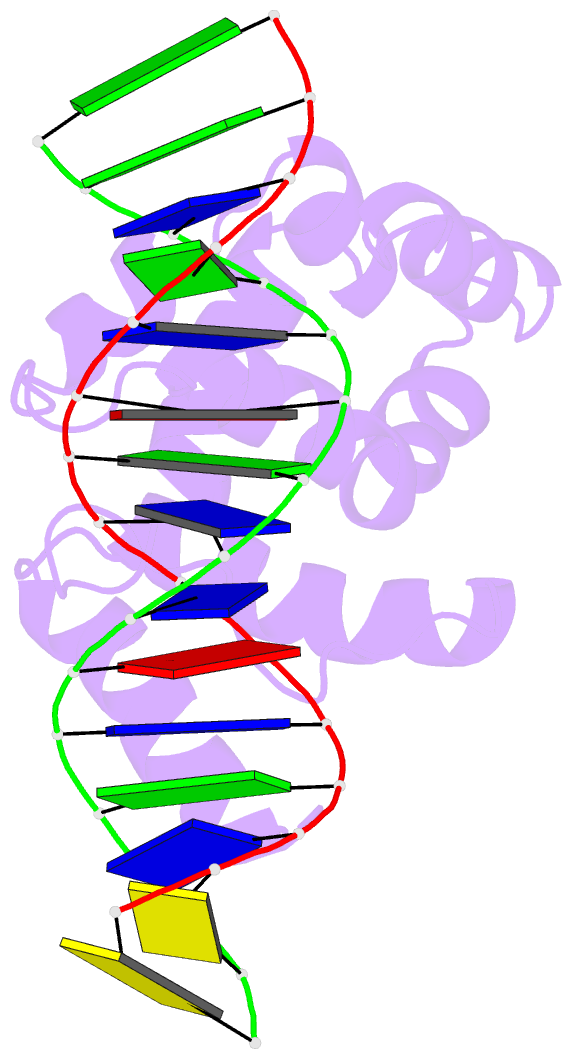
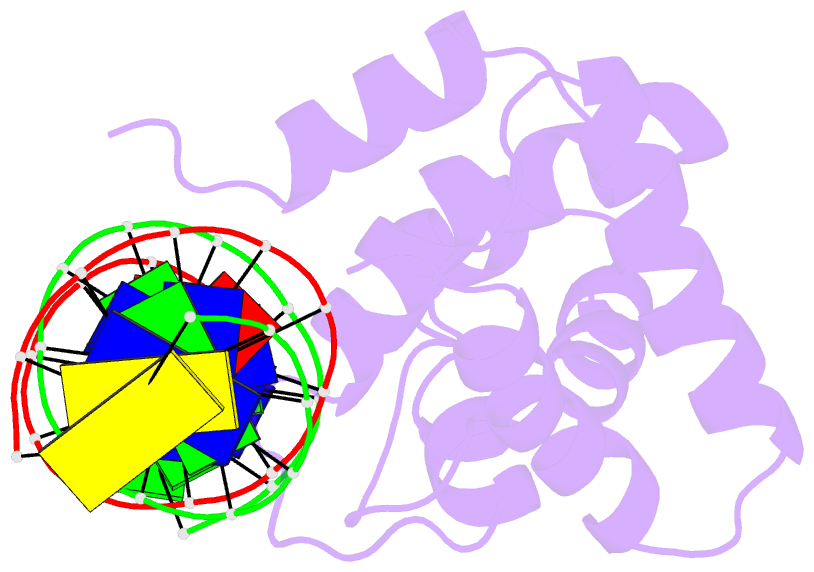
Summary information and primary citation
- PDB-id
- 1kqq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- NMR
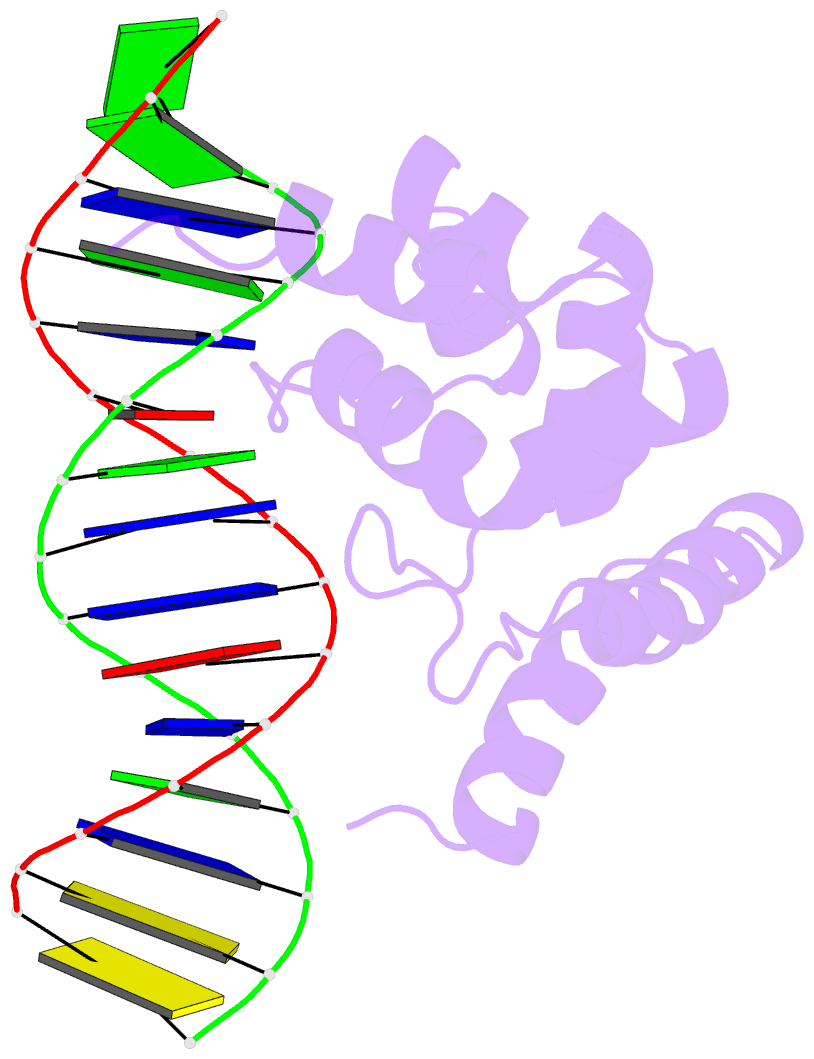
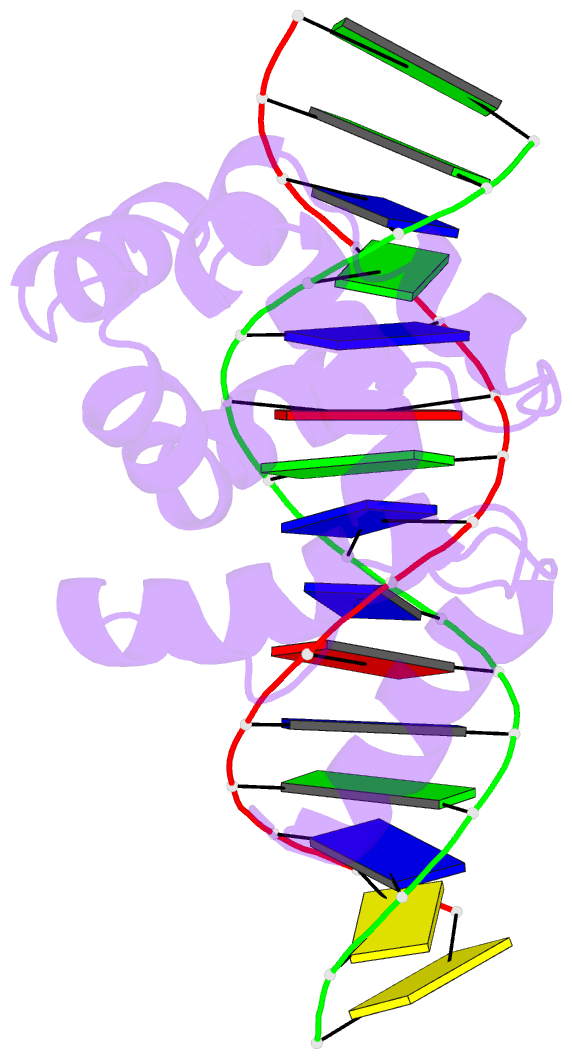
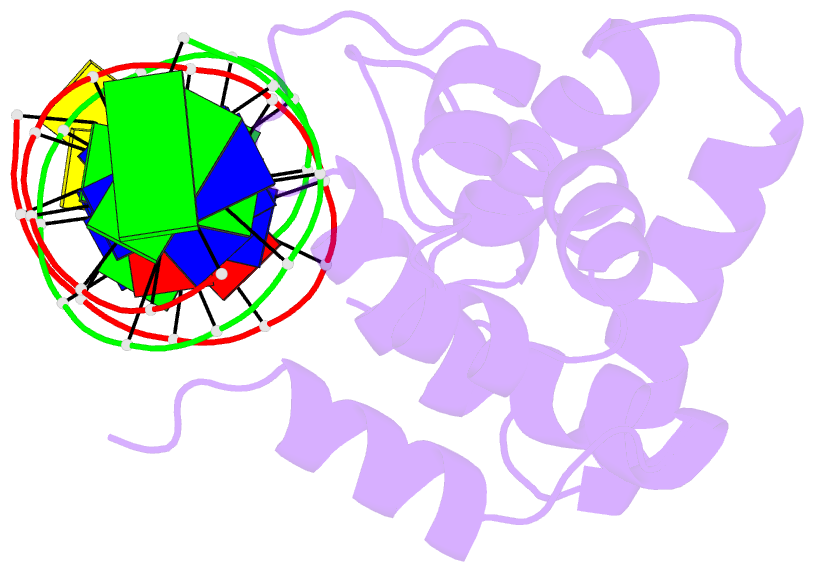
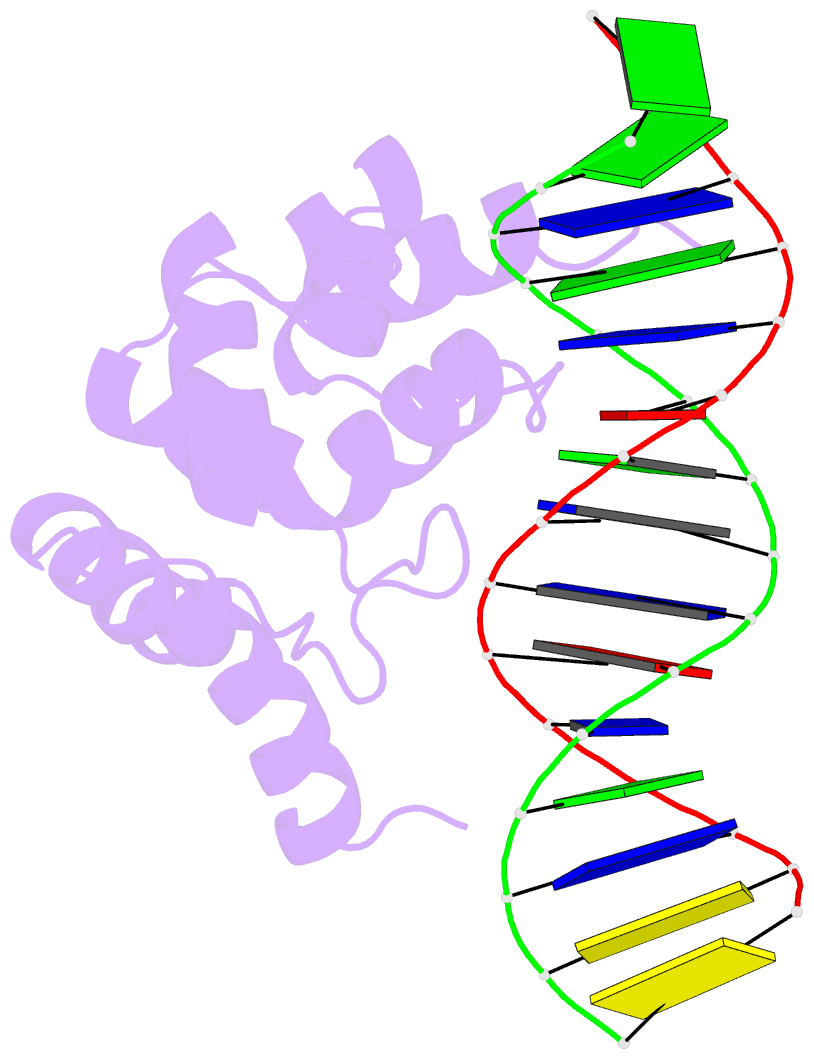
- Summary
- Solution structure of the dead ringer arid-DNA complex
- Reference
- Iwahara J, Iwahara M, Daughdrill GW, Ford J, Clubb RT (2002): "The structure of the Dead ringer-DNA complex reveals how AT-rich interaction domains (ARIDs) recognize DNA." EMBO J., 21, 1197-1209. doi: 10.1093/emboj/21.5.1197.
- Abstract
- The AT-rich interaction domain (ARID) is a DNA-binding module found in many eukaryotic transcription factors. Using NMR spectroscopy, we have determined the first ever three-dimensional structure of an ARID--DNA complex (mol. wt 25.7 kDa) formed by Dead ringer from Drosophila melanogaster. ARIDs recognize DNA through a novel mechanism involving major groove immobilization of a large loop that connects the helices of a non-canonical helix-turn-helix motif, and through a concomitant structural rearrangement that produces stabilizing contacts from a beta-hairpin. Dead ringer's preference for AT-rich DNA originates from three positions within the ARID fold that form energetically significant contacts to an adenine-thymine base step. Amino acids that dictate binding specificity are not highly conserved, suggesting that ARIDs will bind to a range of nucleotide sequences. Extended ARIDs, found in several sequence-specific transcription factors, are distinguished by the presence of a C-terminal helix that may increase their intrinsic affinity for DNA. The prevalence of serine amino acids at all specificity determining positions suggests that ARIDs within SWI/SNF-related complexes will interact with DNA non-sequence specifically.