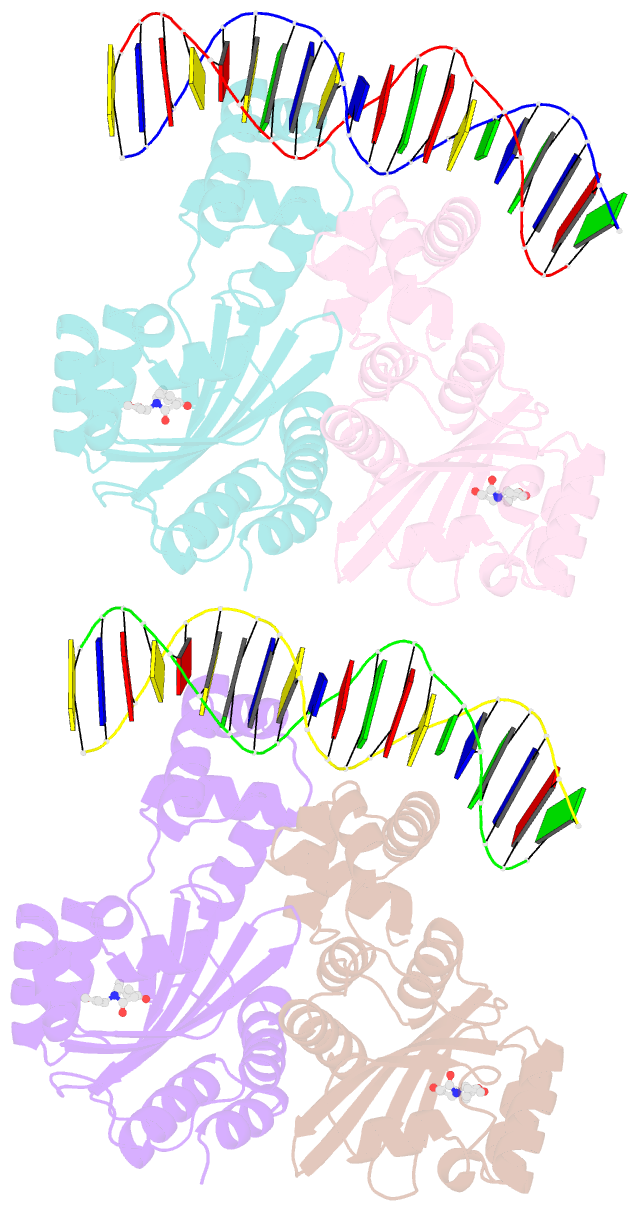
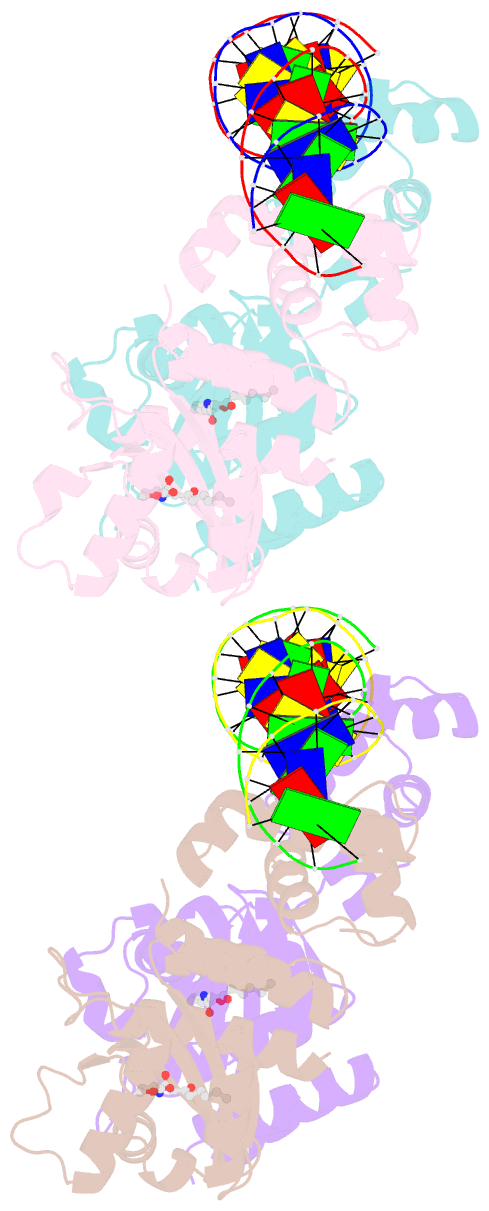
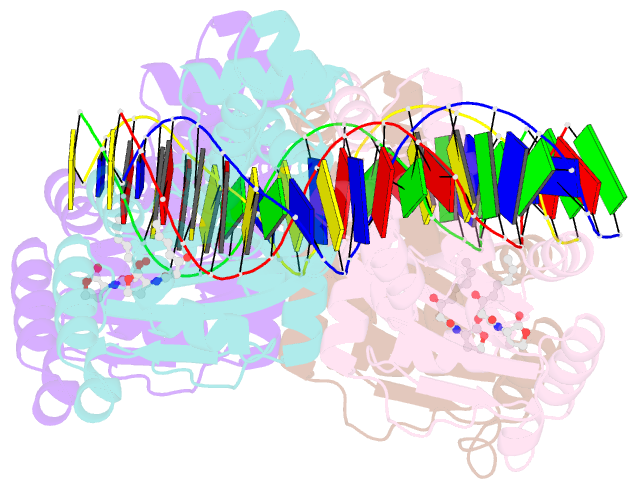
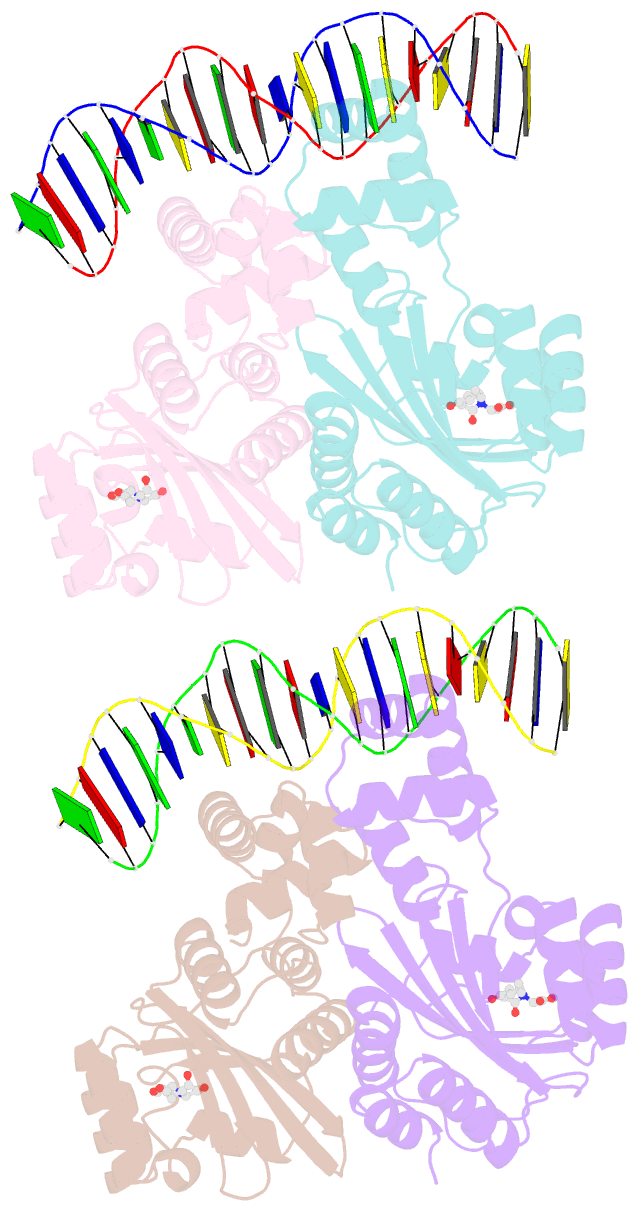
Summary information and primary citation
- PDB-id
- 1l3l; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (1.66 Å)
- Summary
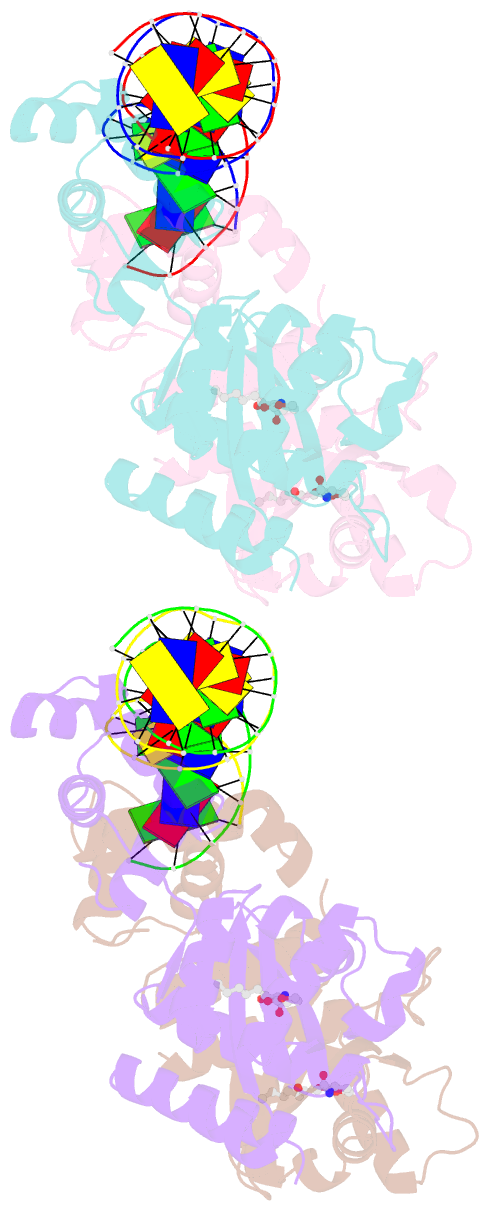
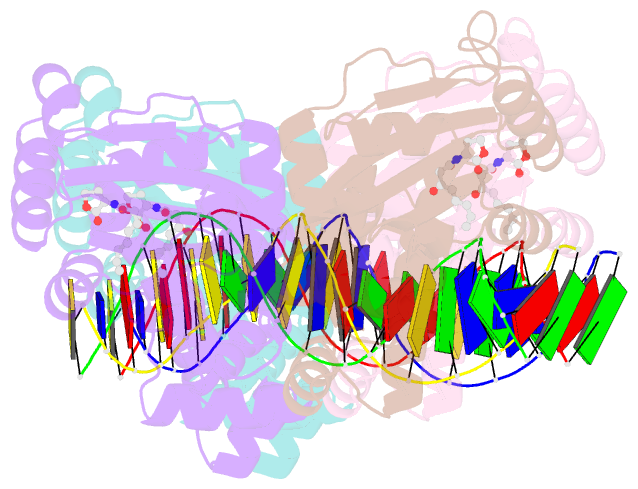
- Crystal structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA
- Reference
- Zhang RG, Pappas T, Brace JL, Miller PC, Oulmassov T, Molyneaux JM, Anderson JC, Bashkin JK, Winans SC, Joachimiak A (2002): "Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA." Nature, 417, 971-974. doi: 10.1038/nature00833.
- Abstract
- Many proteobacteria are able to monitor their population densities through the release of pheromones known as N-acylhomoserine lactones. At high population densities, these pheromones elicit diverse responses that include bioluminescence, biofilm formation, production of antimicrobials, DNA exchange, pathogenesis and symbiosis. Many of these regulatory systems require a pheromone-dependent transcription factor similar to the LuxR protein of Vibrio fischeri. Here we present the structure of a LuxR-type protein. TraR of Agrobacterium tumefaciens was solved at 1.66 A as a complex with the pheromone N-3-oxooctanoyl-L-homoserine lactone (OOHL) and its TraR DNA-binding site. The amino-terminal domain of TraR is an alpha/beta/alpha sandwich that binds OOHL, whereas the carboxy-terminal domain contains a helix turn helix DNA-binding motif. The TraR dimer displays a two-fold symmetry axis in each domain; however, these two axes of symmetry are at an approximately 90 degree angle, resulting in a pronounced overall asymmetry of the complex. The pheromone lies fully embedded within the protein with virtually no solvent contact, and makes numerous hydrophobic contacts with the protein as well as four hydrogen bonds: three direct and one water-mediated.