Summary information and primary citation
- PDB-id
- 1lng; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- signaling protein-RNA
- Method
- X-ray (2.3 Å)
- Summary
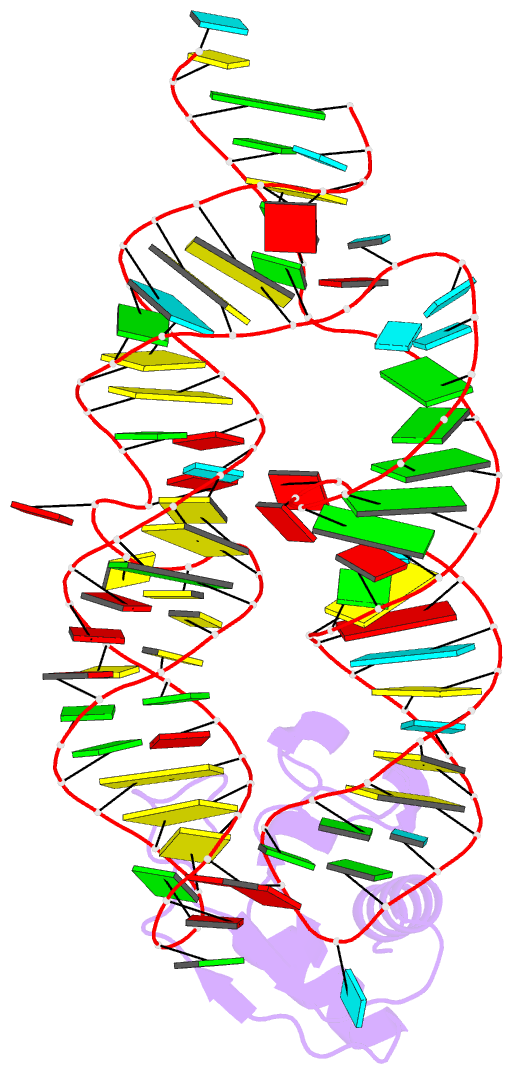
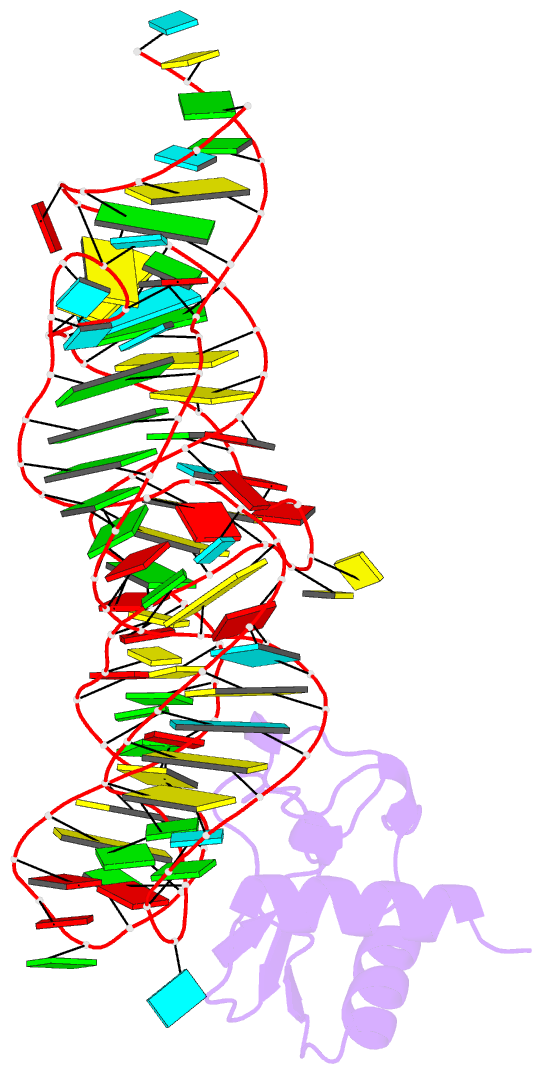
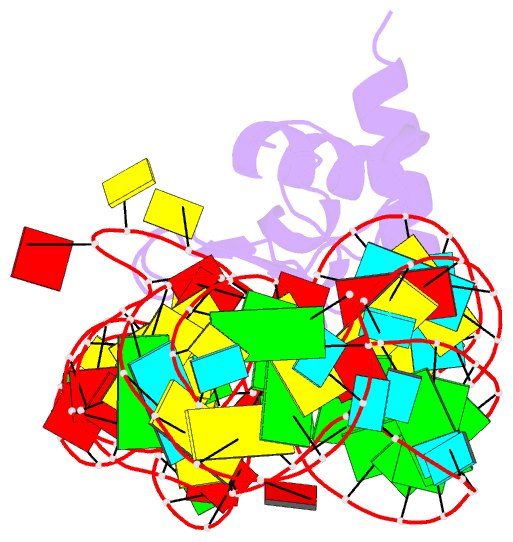
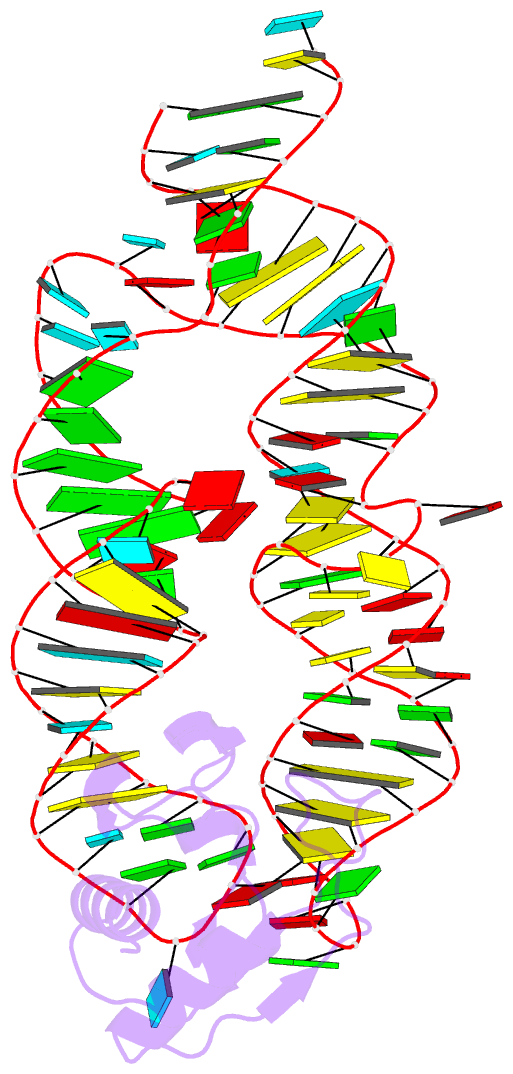
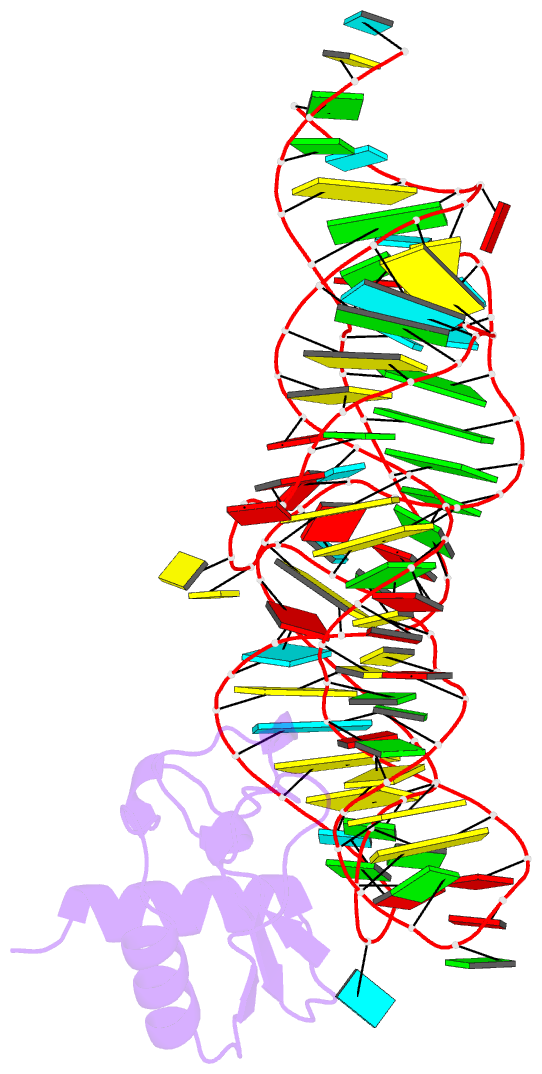
- Crystal structure of the srp19-7s.s srp RNA complex of m. jannaschii
- Reference
- Hainzl T, Huang S, Sauer-Eriksson AE (2002): "Structure of the SRP19 RNA complex and implications for signal recognition particle assembly." Nature, 417, 767-771. doi: 10.1038/nature00768.
- Abstract
- The signal recognition particle (SRP) is a phylogenetically conserved ribonucleoprotein. It associates with ribosomes to mediate co-translational targeting of membrane and secretory proteins to biological membranes. In mammalian cells, the SRP consists of a 7S RNA and six protein components. The S domain of SRP comprises the 7S.S part of RNA bound to SRP19, SRP54 and the SRP68/72 heterodimer; SRP54 has the main role in recognizing signal sequences of nascent polypeptide chains and docking SRP to its receptor. During assembly of the SRP, binding of SRP19 precedes and promotes the association of SRP54 (refs 4, 5). Here we report the crystal structure at 2.3 A resolution of the complex formed between 7S.S RNA and SRP19 in the archaeon Methanococcus jannaschii. SRP19 bridges the tips of helices 6 and 8 of 7S.S RNA by forming an extensive network of direct protein RNA interactions. Helices 6 and 8 pack side by side; tertiary RNA interactions, which also involve the strictly conserved tetraloop bases, stabilize helix 8 in a conformation competent for SRP54 binding. The structure explains the role of SRP19 and provides a molecular framework for SRP54 binding and SRP assembly in Eukarya and Archaea.