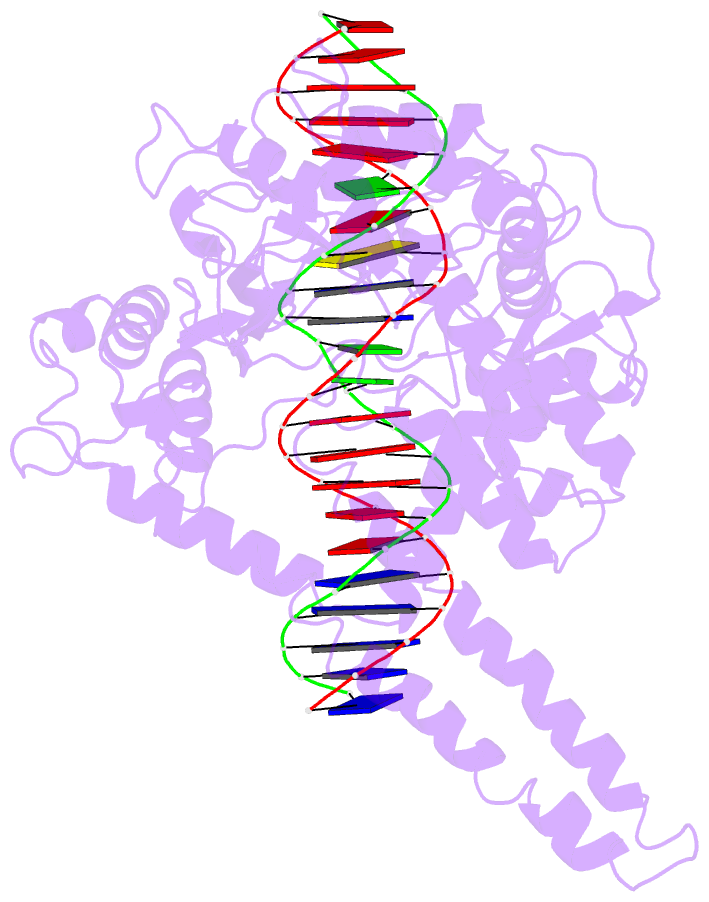
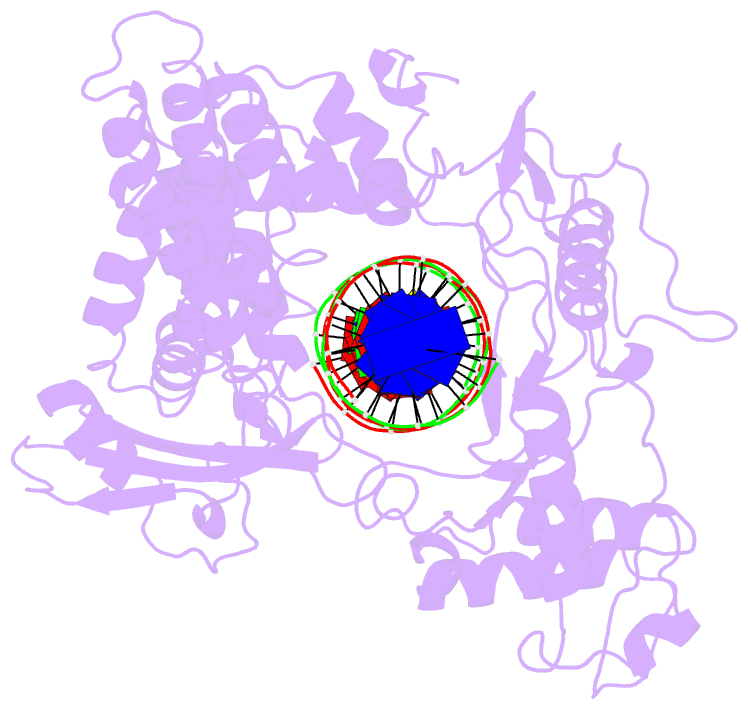
Summary information and primary citation
- PDB-id
- 1lpq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- isomerase-DNA
- Method
- X-ray (3.14 Å)
- Summary
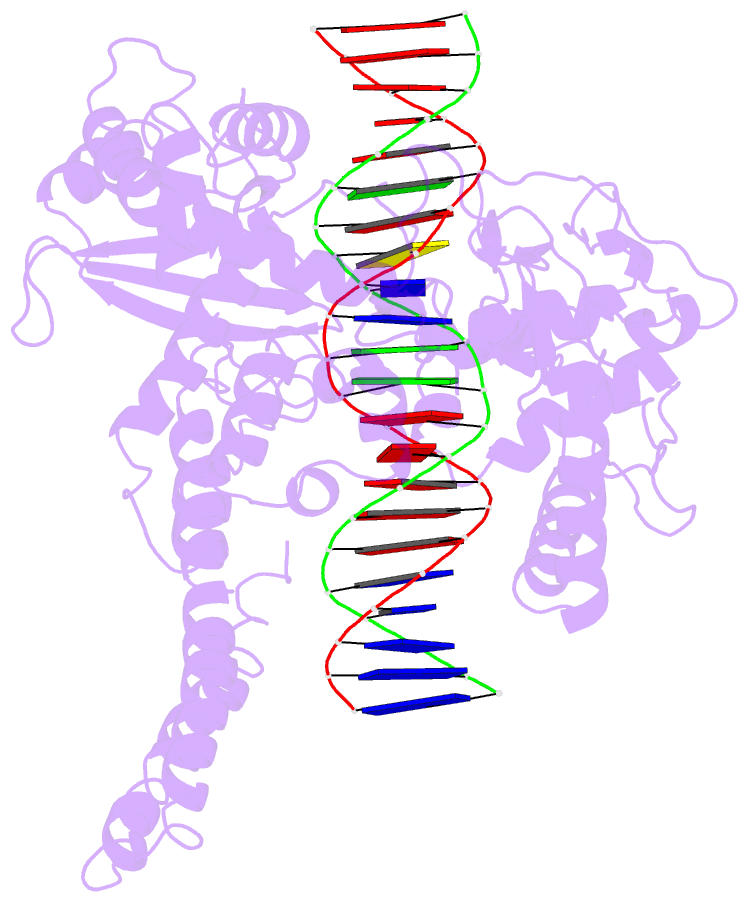
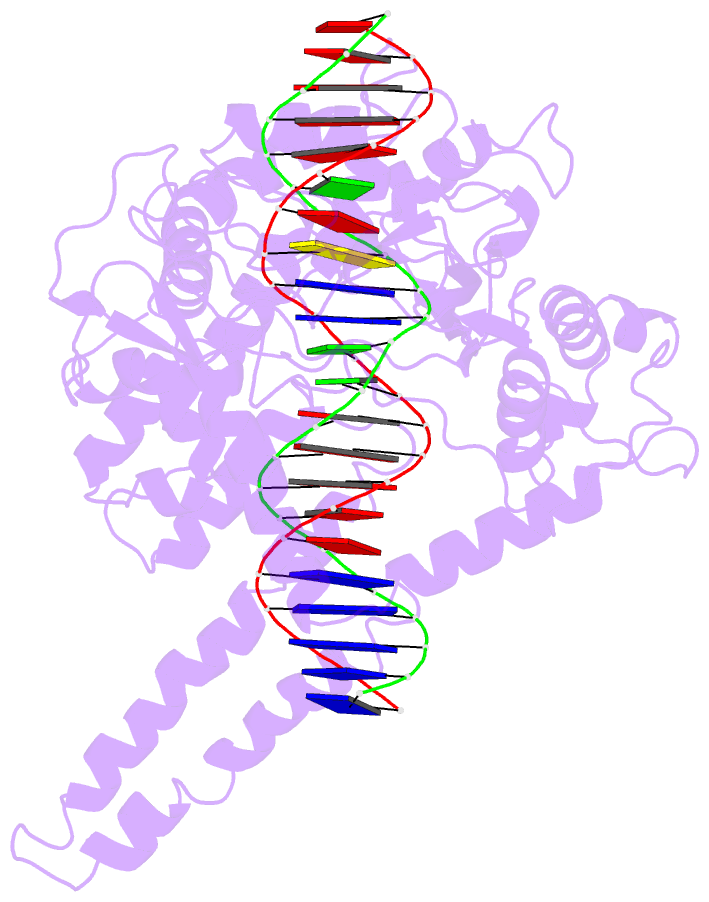
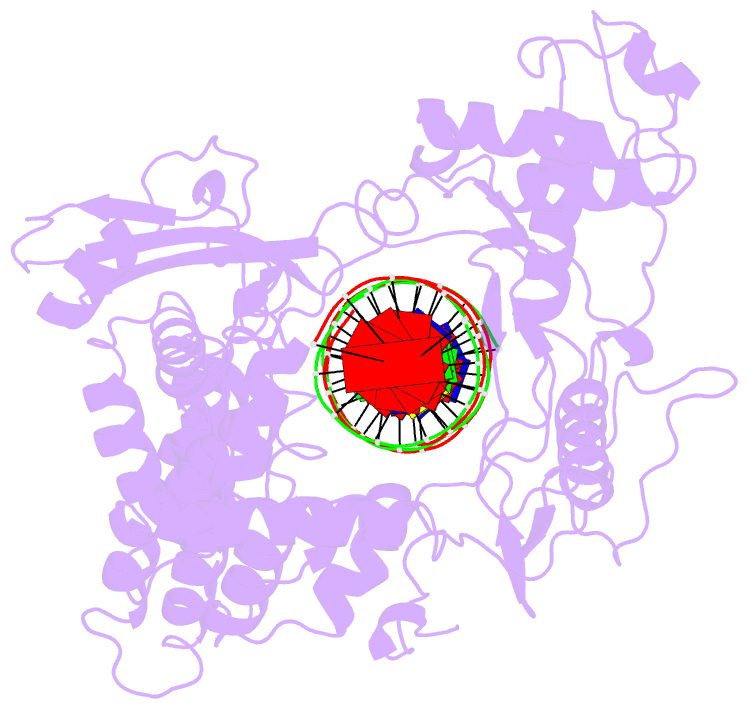
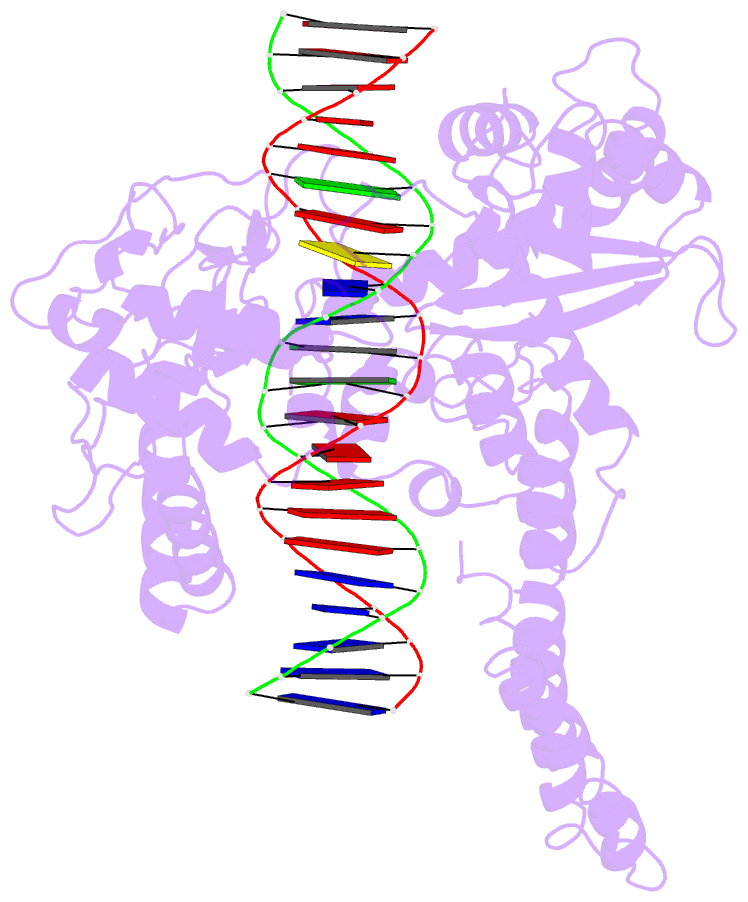
- Human DNA topoisomerase i (70 kda) in non-covalent complex with a 22 base pair DNA duplex containing an 8-oxog lesion
- Reference
- Lesher DT, Pommier Y, Stewart L, Redinbo MR (2002): "8-Oxoguanine rearranges the active site of human topoisomerase I." Proc.Natl.Acad.Sci.USA, 99, 12102-12107. doi: 10.1073/pnas.192282699.
- Abstract
- 7,8-Dihydro-8-oxoguanine (8-oxoG) is the most common form of oxidative DNA damage in human cells. Biochemical studies have shown that 8-oxoG decreases the DNA cleavage activity of human topoisomerase I, an enzyme vital to DNA metabolism and stability. We present the 3.1-A crystal structure of human topoisomerase I in noncovalent complex with a DNA oligonucleotide containing 8-oxoG at the +1 position in the scissile strand. We find that 8-oxoG reorganizes the active site of human topoisomerase I into an inactive conformation relative to the structures of topoisomerase I-DNA complexes elucidated previously. The catalytic Tyr-723-Phe rotates away from the DNA cleavage site and packs into the body of the molecule. A second active-site residue, Arg-590, becomes disordered and is not observed in the structure. The docked, inactive conformation of Tyr-723-Phe is reminiscent of the related tyrosine recombinase family of integrases and recombinases, suggesting a common regulatory mechanism. We propose that human topoisomerase I binds to DNA first in an inactive conformation and then rearranges its active site for catalysis. 8-OxoG appears to impact topoisomerase I by stabilizing the inactive, DNA-bound state.