Summary information and primary citation
- PDB-id
- 1m8v; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (2.6 Å)
- Summary
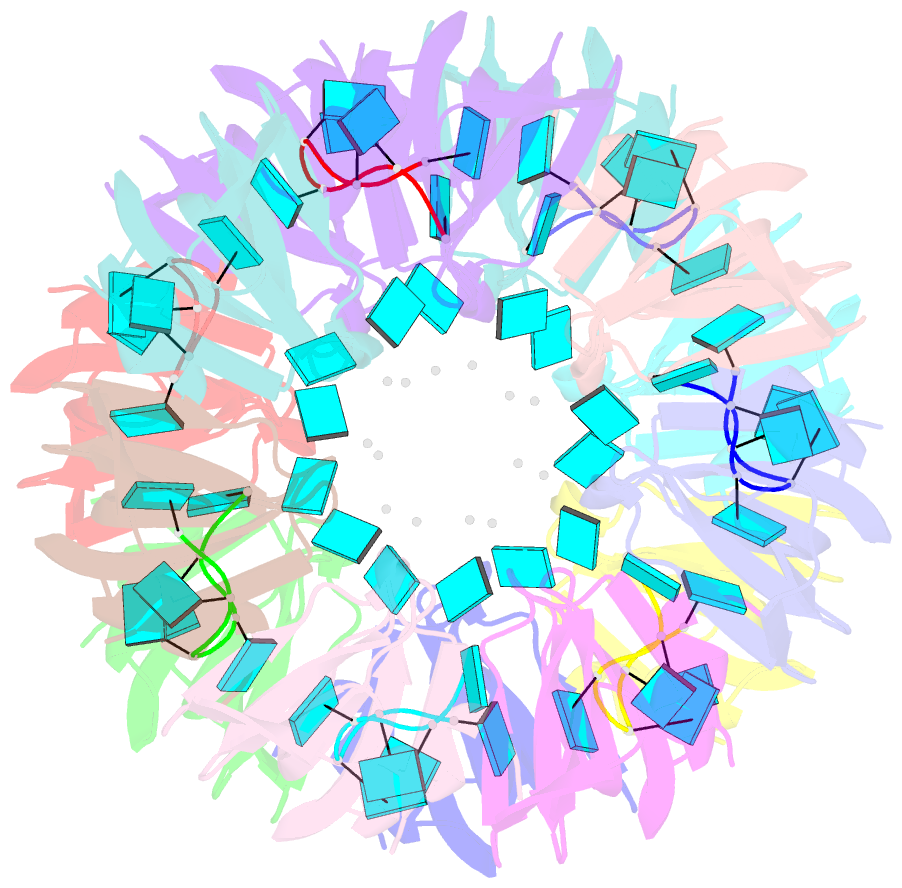
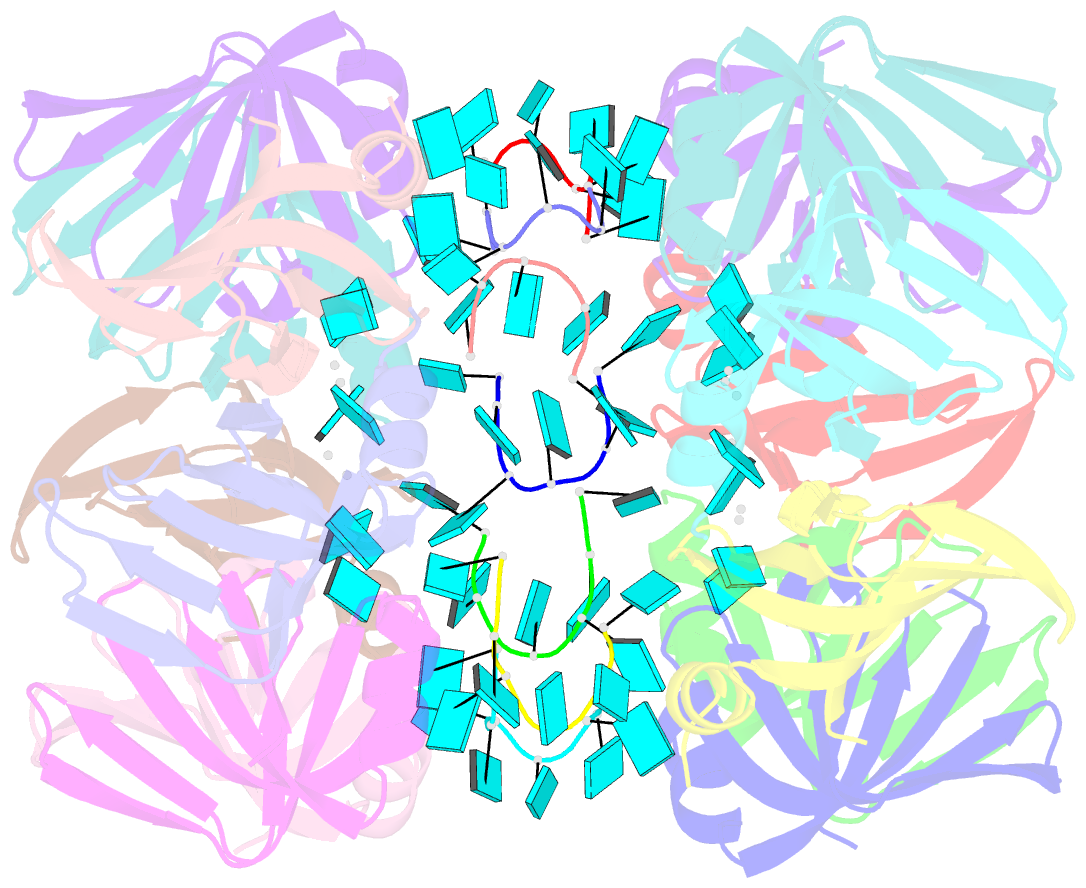
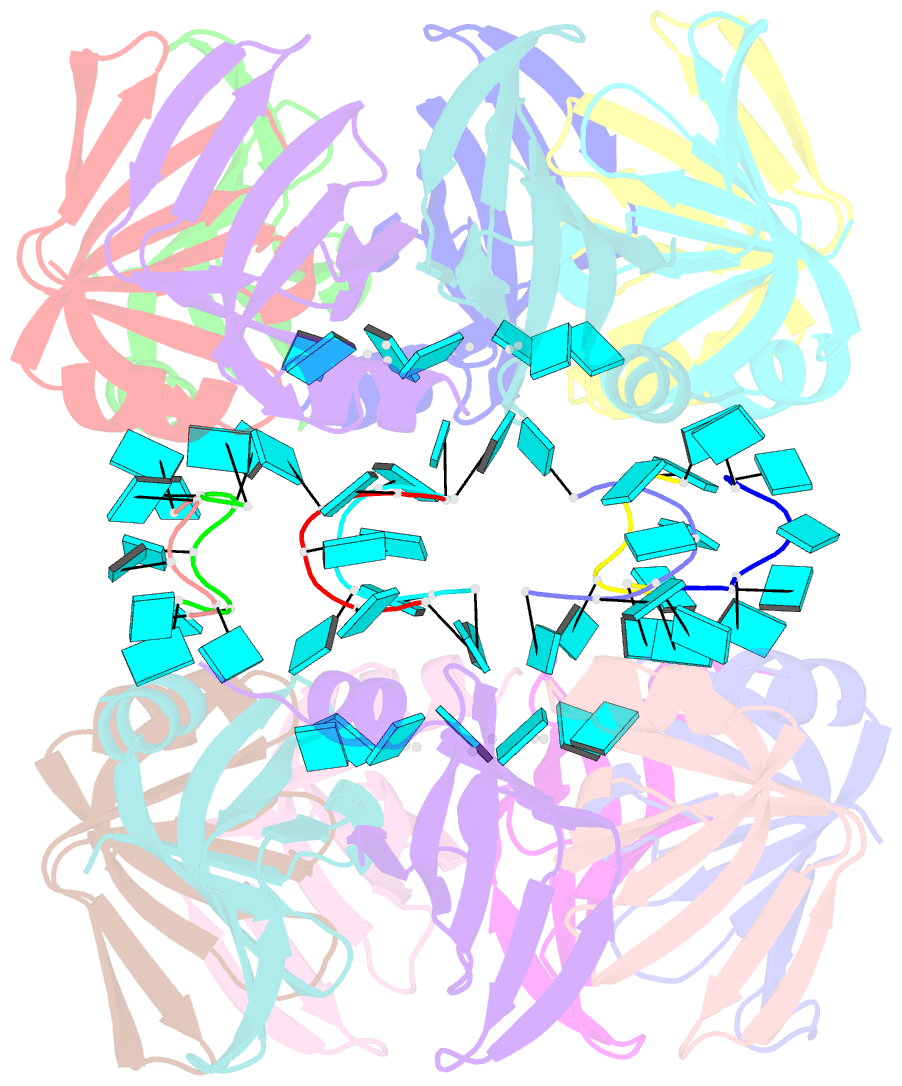
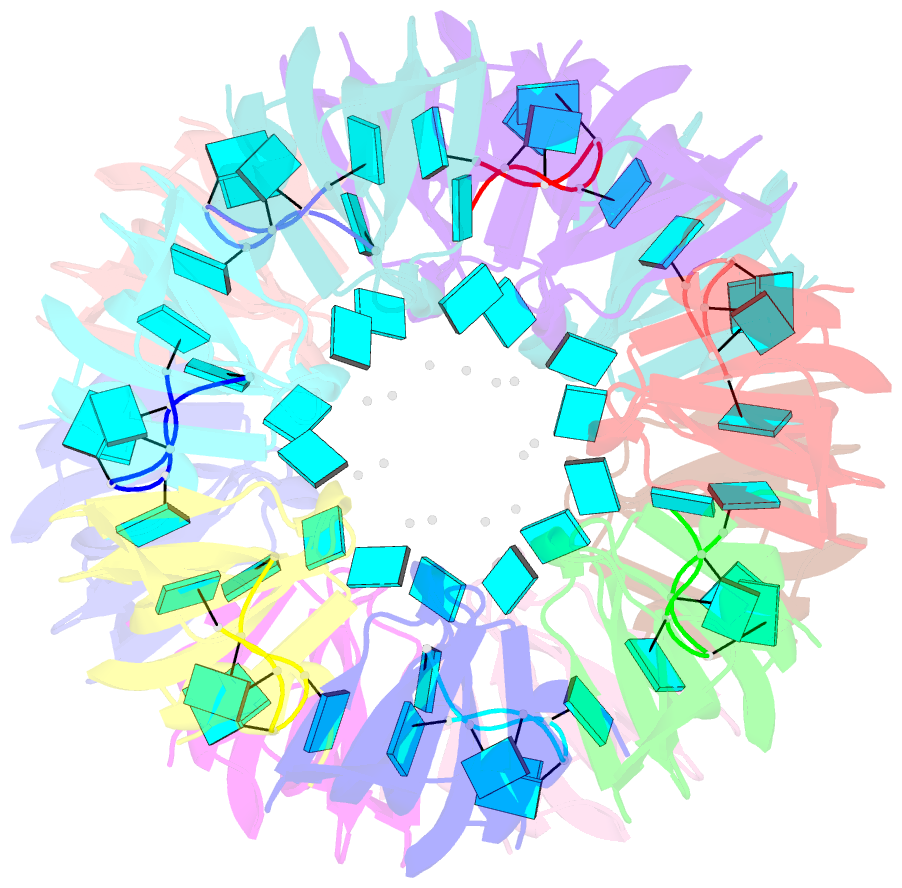
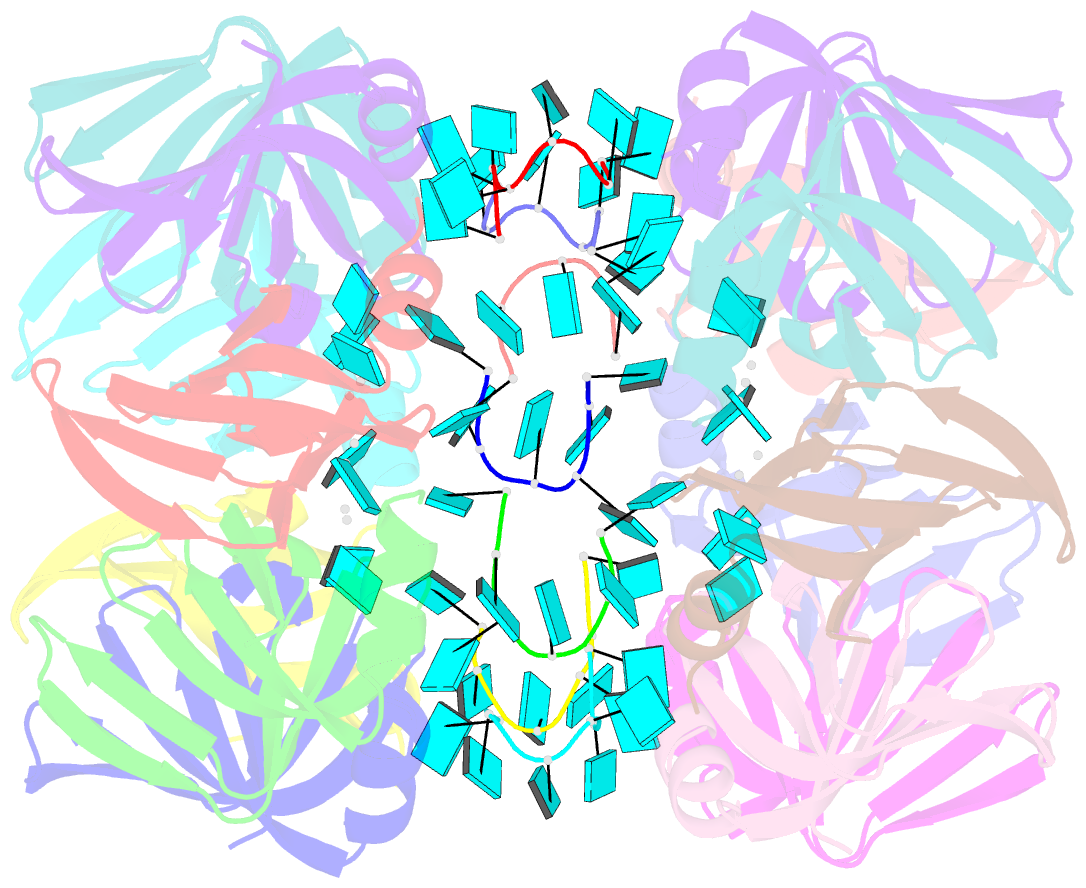
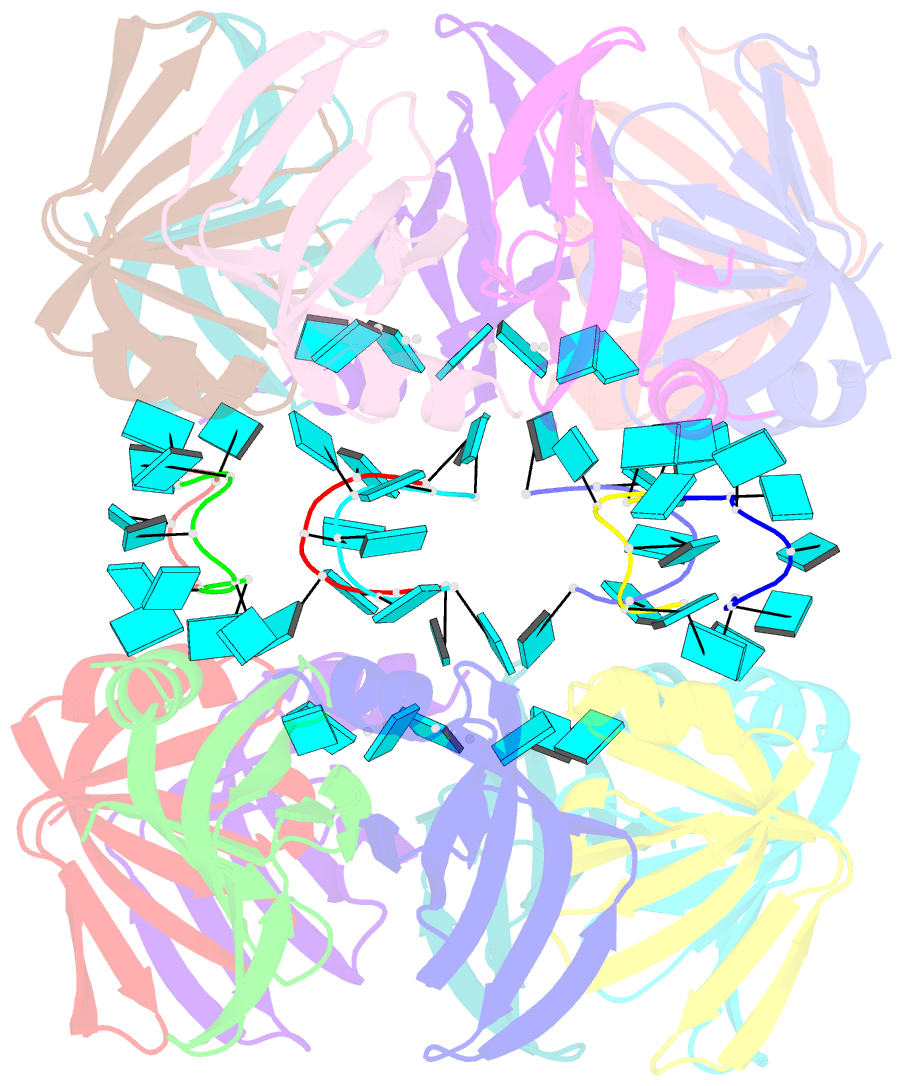
- Structure of pyrococcus abyssii sm protein in complex with a uridine heptamer
- Reference
- Thore S, Mayer C, Sauter C, Weeks S, Suck D (2003): "Crystal Structure of Pyrococcus abyssii Sm core and its Complex with RNA: Common Features of RNA-binding in Archaea and Eukarya." J.Biol.Chem., 278, 1239-1247. doi: 10.1074/jbc.M207685200.
- Abstract
- The Sm proteins are conserved in all three domains of life and are always associated with U-rich RNA sequences. Their proposed function is to mediate RNA-RNA interactions. We present here the crystal structures of Pyrococcus abyssi Sm protein (PA-Sm1) and its complex with a uridine heptamer. The overall structure of the protein complex, a heptameric ring with a central cavity, is similar to that proposed for the eukaryotic Sm core complex and found for other archaeal Sm proteins. RNA molecules bind to the protein at two different sites. They interact specifically inside the ring with three highly conserved residues, defining the uridine-binding pocket. In addition, nucleotides also interact on the surface formed by the N-terminal alpha-helix as well as a conserved aromatic residue in beta-strand 2 of the PA-Sm1 protein. The mutation of this conserved aromatic residue shows the importance of this second site for the discrimination between RNA sequences. Given the high structural homology between archaeal and eukaryotic Sm proteins, the PA-Sm1.RNA complex provides a model for how the small nuclear RNA contacts the Sm proteins in the Sm core. In addition, it suggests how Sm proteins might exert their function as modulators of RNA-RNA interactions.