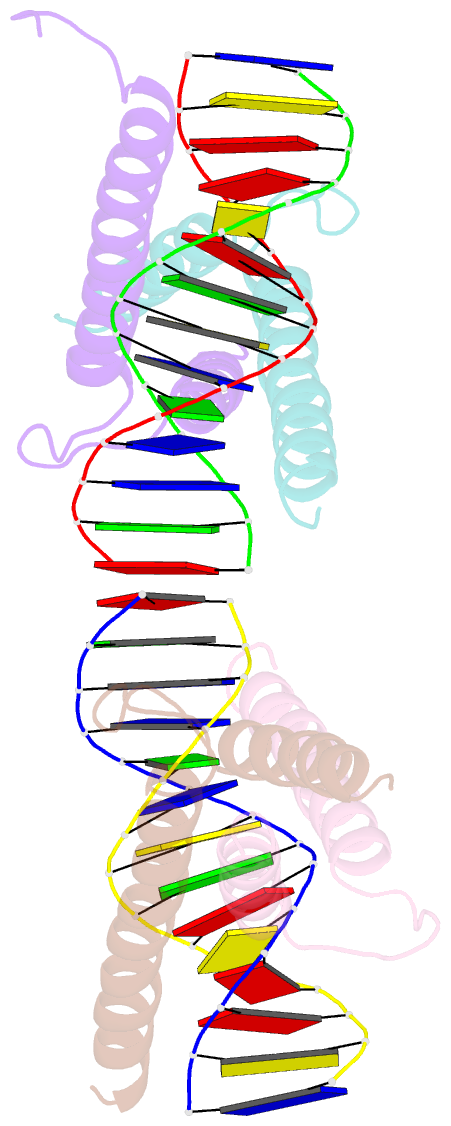
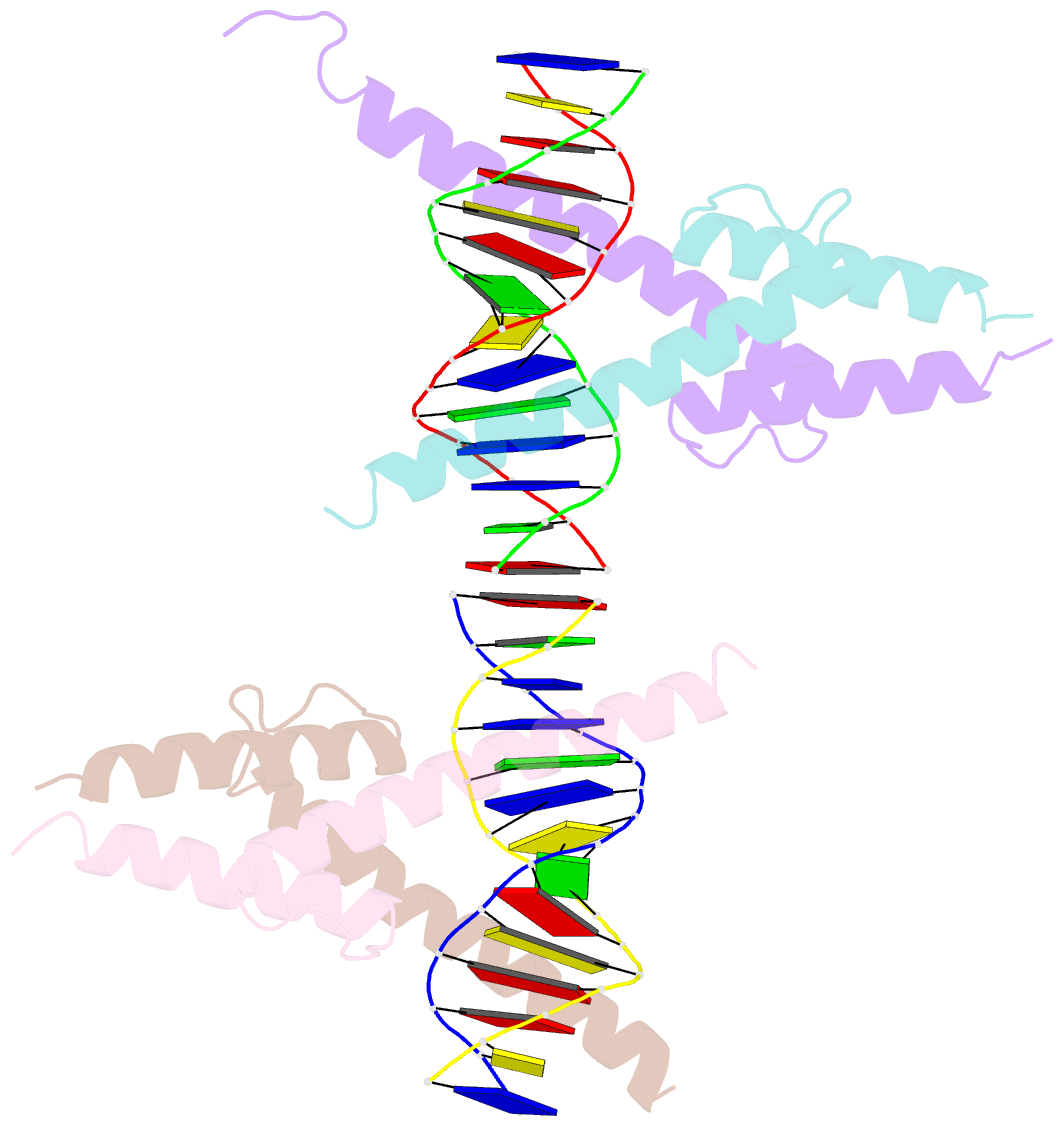
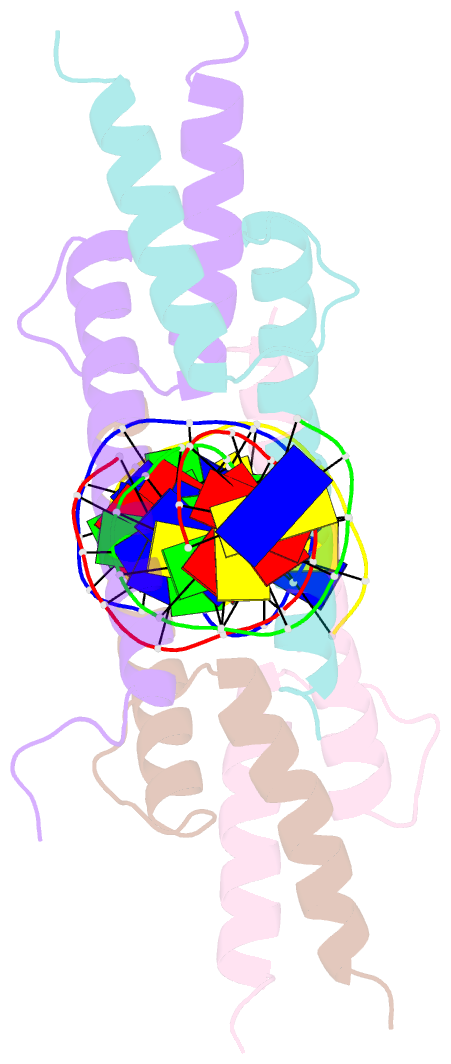
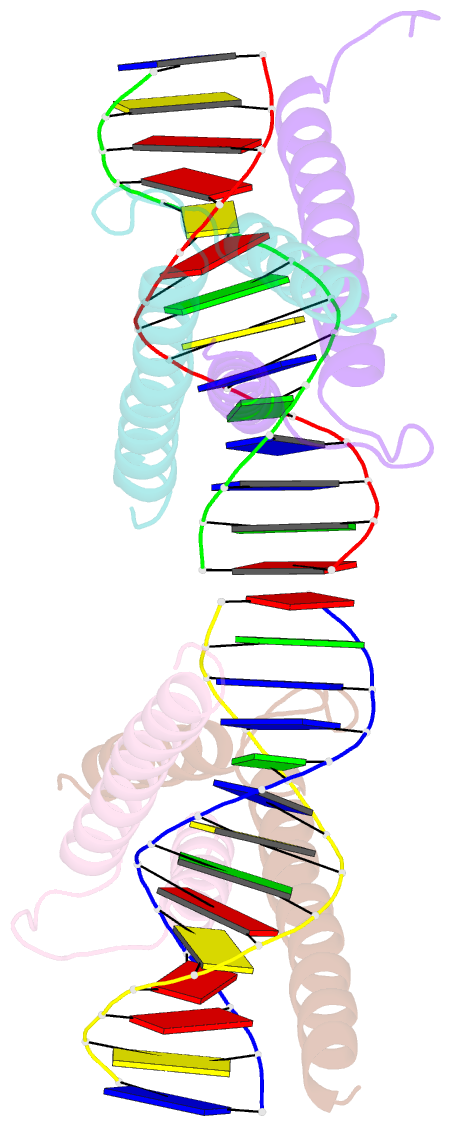
Summary information and primary citation
- PDB-id
- 1mdy; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.8 Å)
- Summary
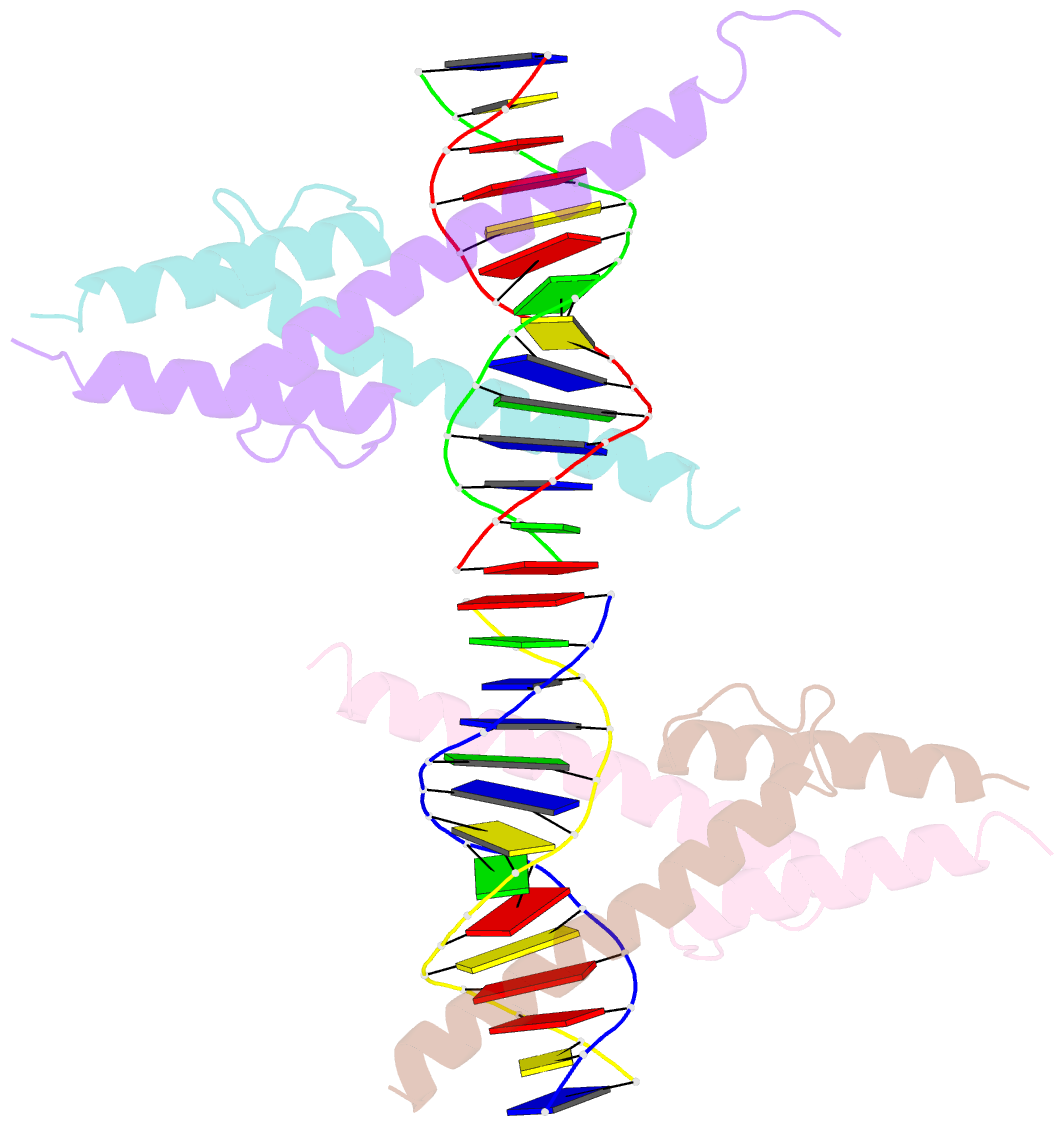
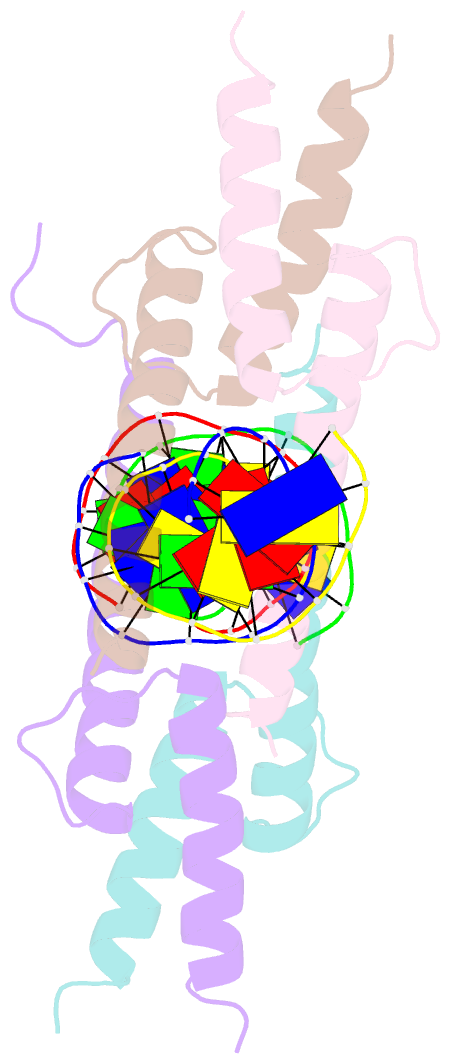
- Crystal structure of myod bhlh domain bound to DNA: perspectives on DNA recognition and implications for transcriptional activation
- Reference
- Ma PC, Rould MA, Weintraub H, Pabo CO (1994): "Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation." Cell(Cambridge,Mass.), 77, 451-459. doi: 10.1016/0092-8674(94)90159-7.
- Abstract
- The crystal structure of a MyoD basic-helix-loop-helix (bHLH) domain-DNA complex has been solved and refined at 2.8 A resolution. This structure proves that bHLH and bHLH-leucine zipper (bHLH-ZIP) proteins are remarkably similar; it helps us understand subtle differences in binding preferences for these proteins; and it has surprising implications for our understanding of transcription. Specifically, Ala-114 and Thr-115, which are required for positive control in the myogenic proteins, are buried at the protein-DNA interface. These residues are not available for direct protein-protein contacts, but they may determine the conformation of Arg-111. Comparisons with Max suggest that the conformation of this arginine, which is different in the two structures, may play an important role in myogenic transcription.