Summary information and primary citation
- PDB-id
- 1mhd; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- complex (transcription activator-DNA)
- Method
- X-ray (2.8 Å)
- Summary
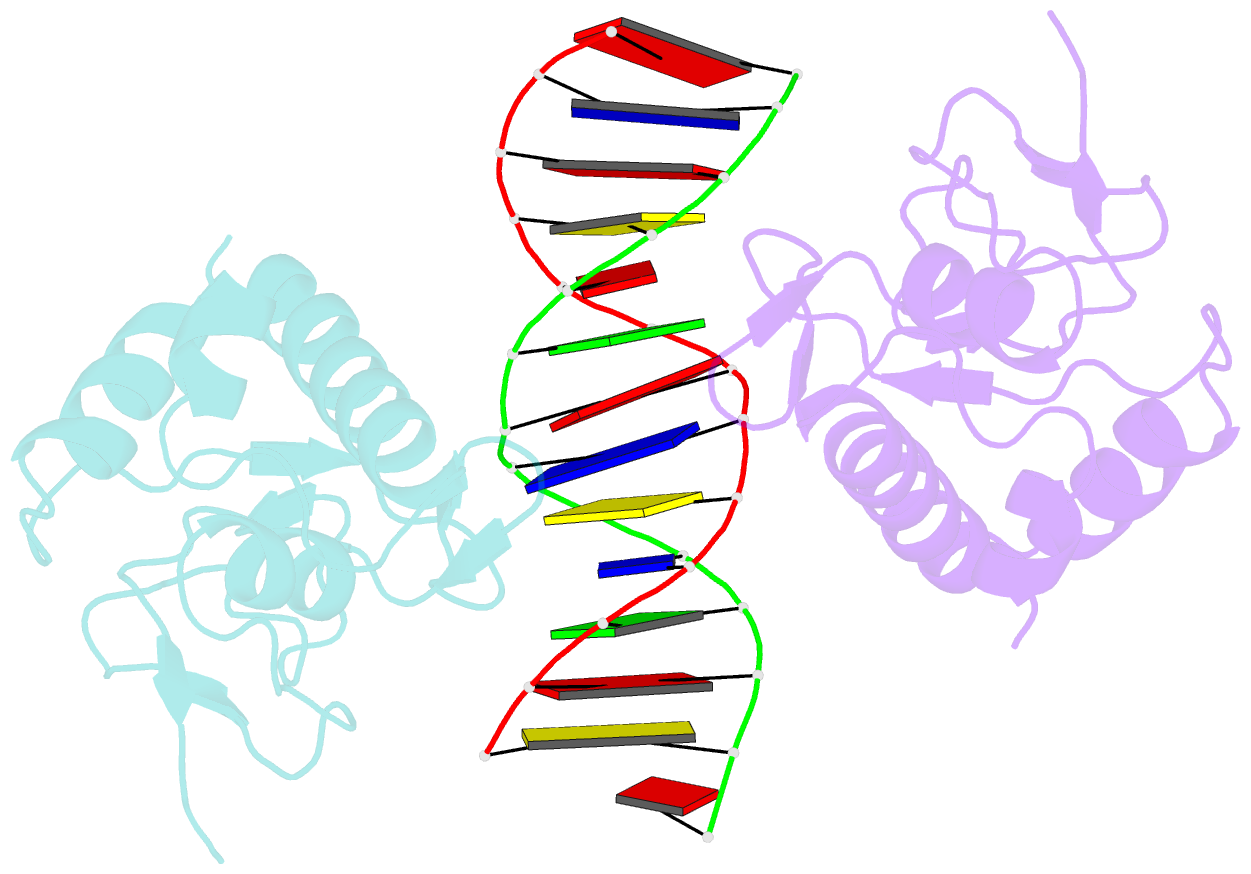
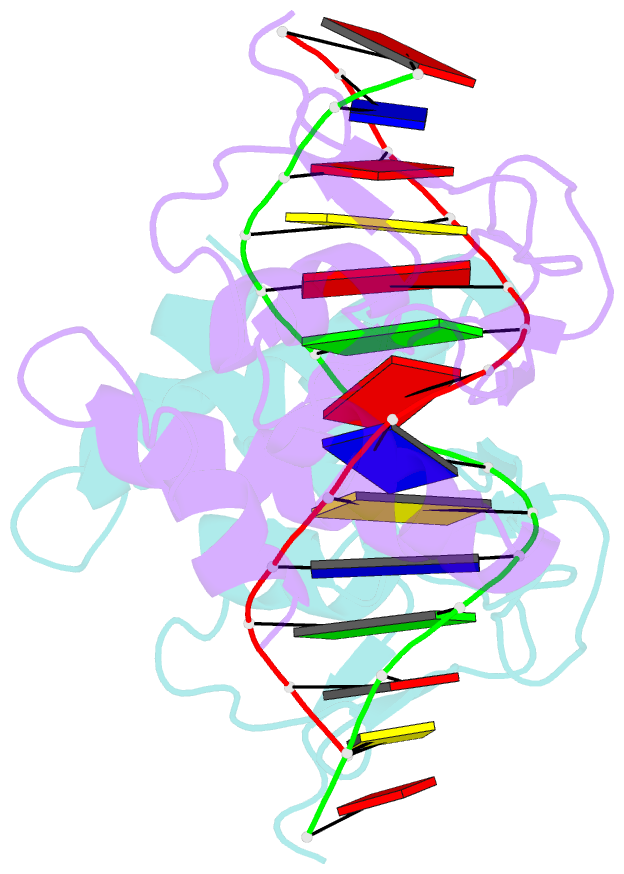
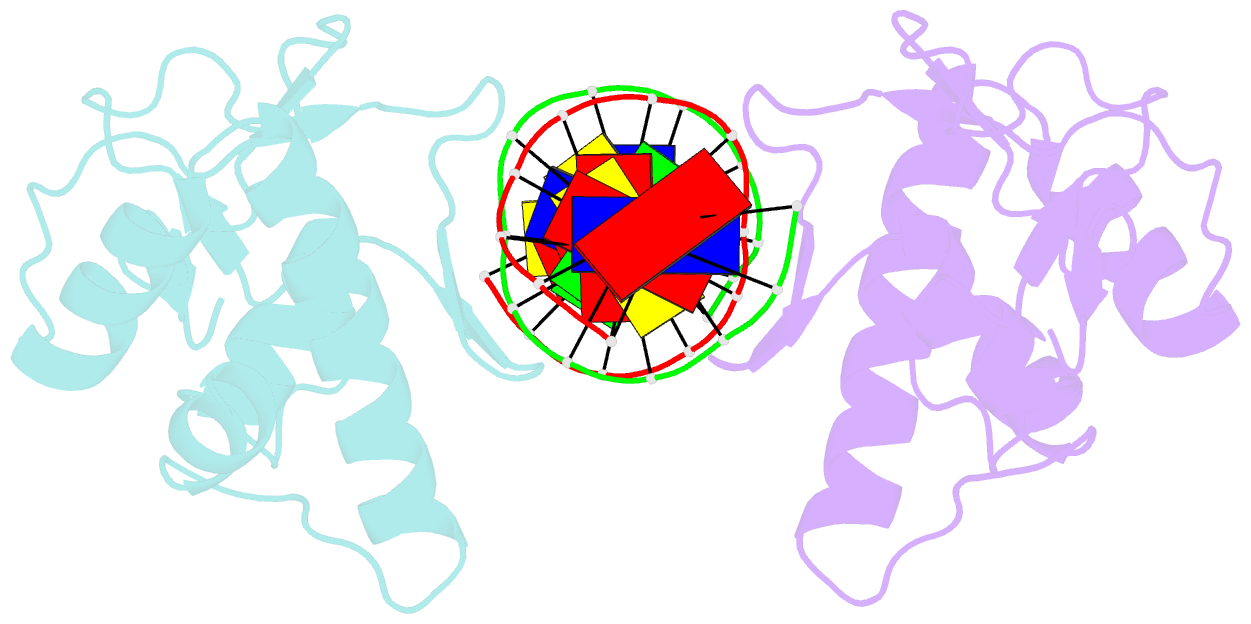
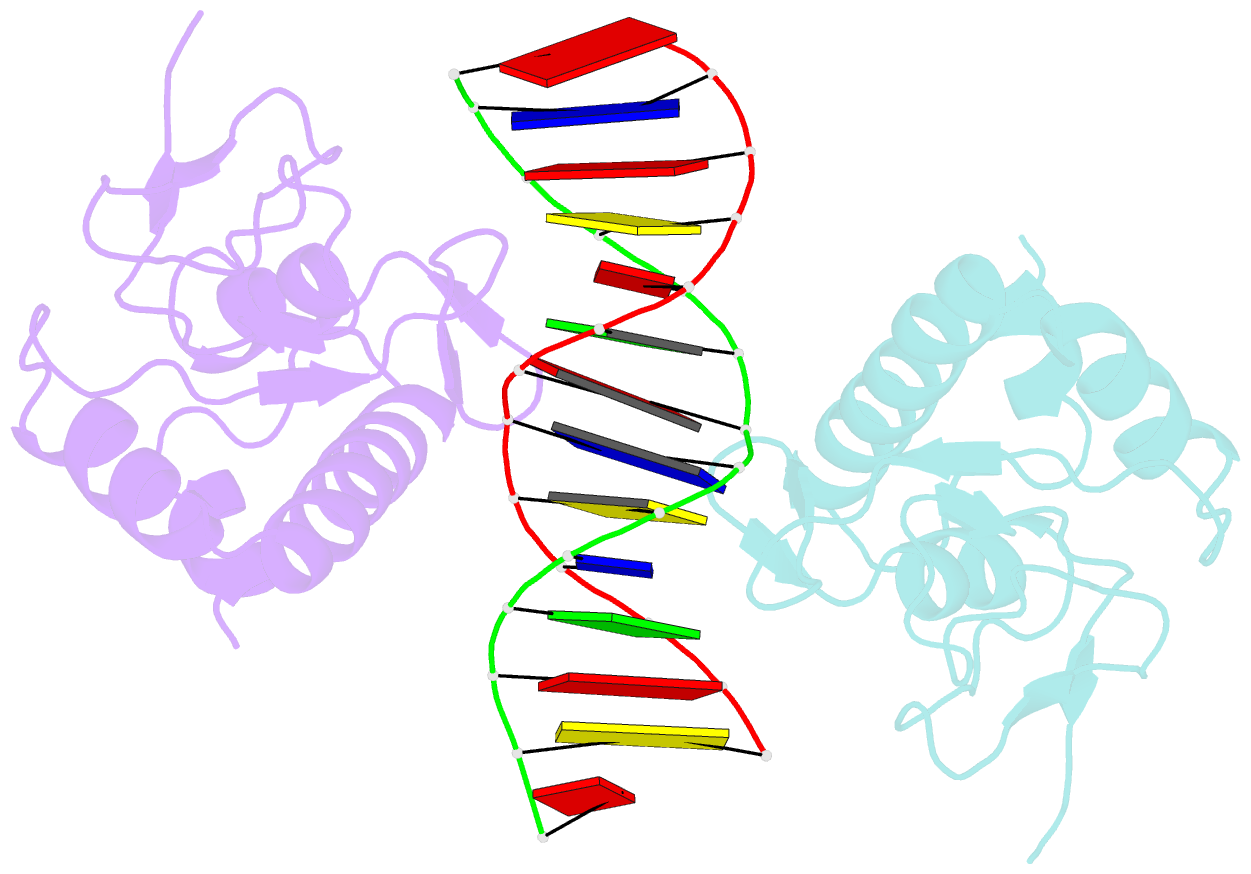
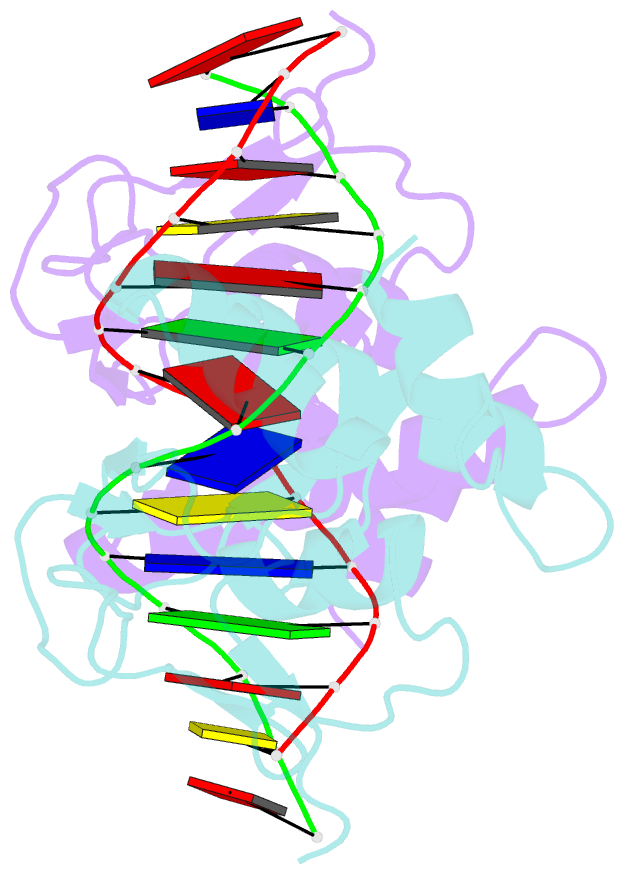
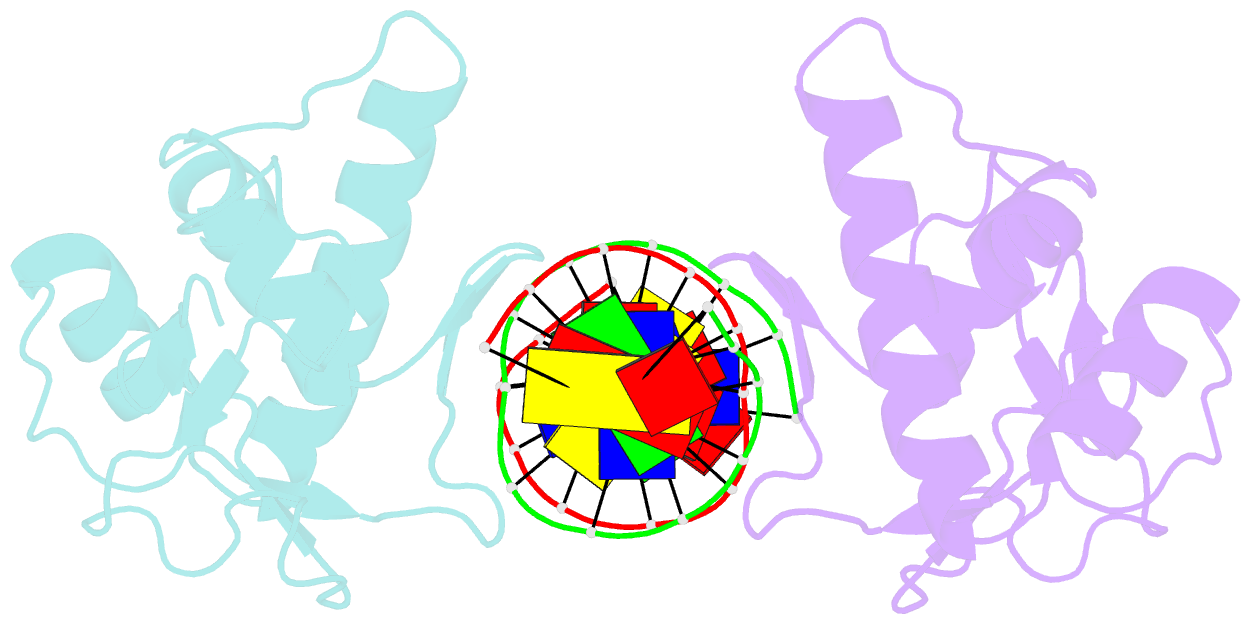
- Crystal structure of a smad mh1 domain bound to DNA
- Reference
- Shi Y, Wang YF, Jayaraman L, Yang H, Massague J, Pavletich NP (1998): "Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling." Cell(Cambridge,Mass.), 94, 585-594. doi: 10.1016/S0092-8674(00)81600-1.
- Abstract
- The Smad family of proteins, which are frequently targeted by tumorigenic mutations in cancer, mediate TGF-beta signaling from cell membrane to nucleus. The crystal structure of a Smad3 MH1 domain bound to an optimal DNA sequence determined at 2.8 A resolution reveals a novel DNA-binding motif. In the crystals, base-specific DNA recognition is provided exclusively by a conserved 11-residue beta hairpin that is embedded in the major groove of DNA. A surface loop region, to which tumorigenic mutations map, has been identified as a functional surface important for Smad activity. This structure establishes a framework for understanding how Smad proteins may act in concert with other transcription factors in the regulation of TGF-beta-responsive genes.