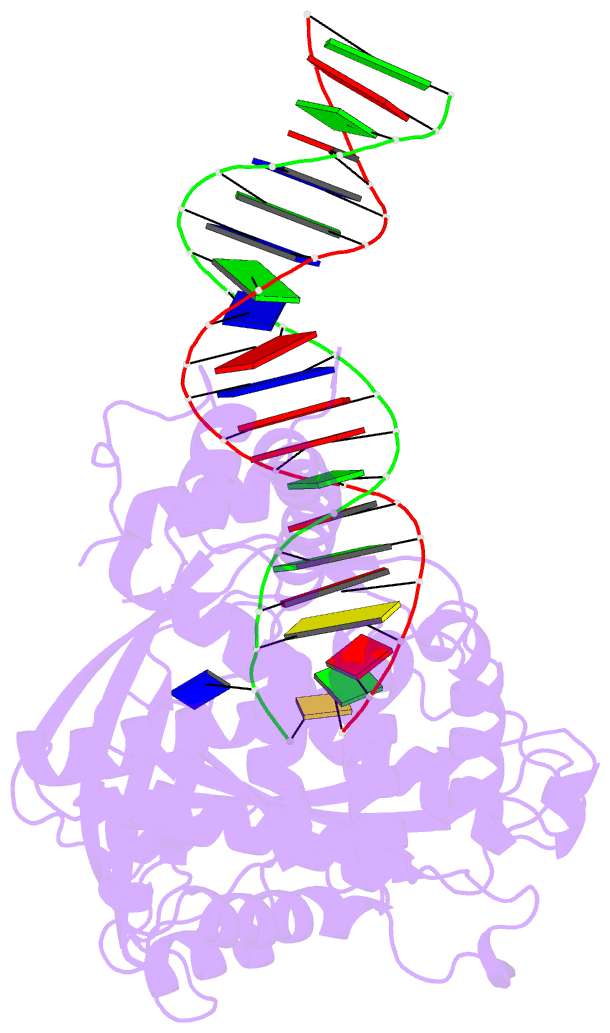
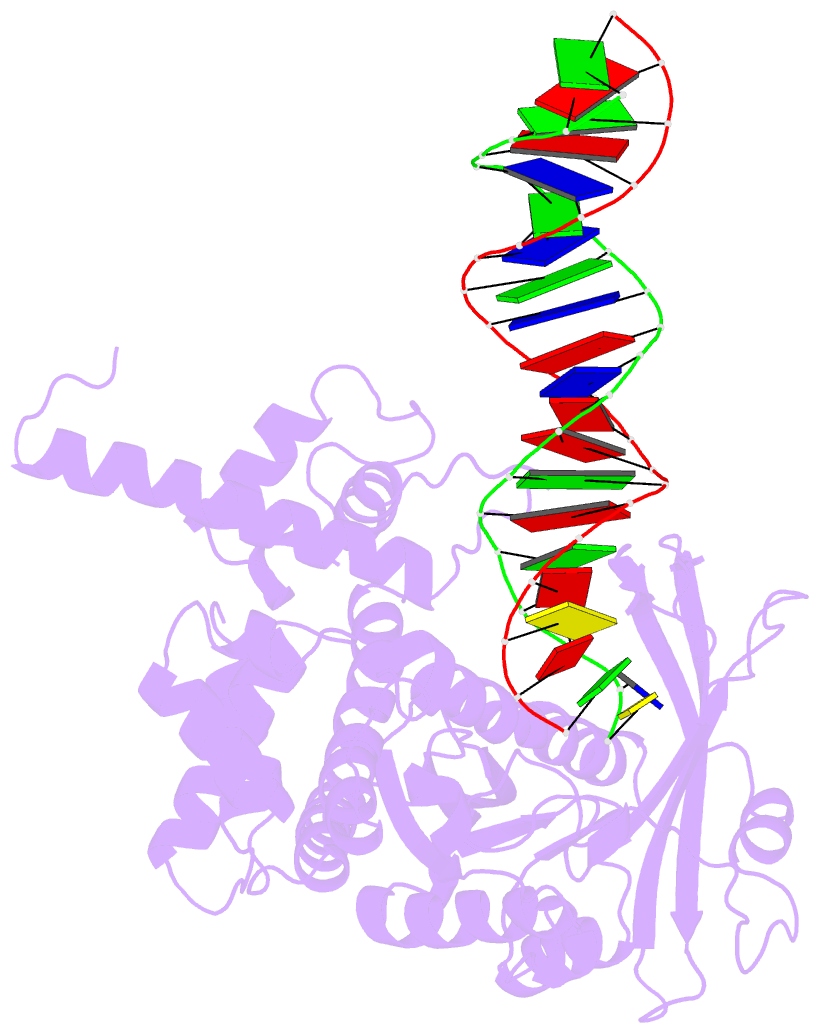
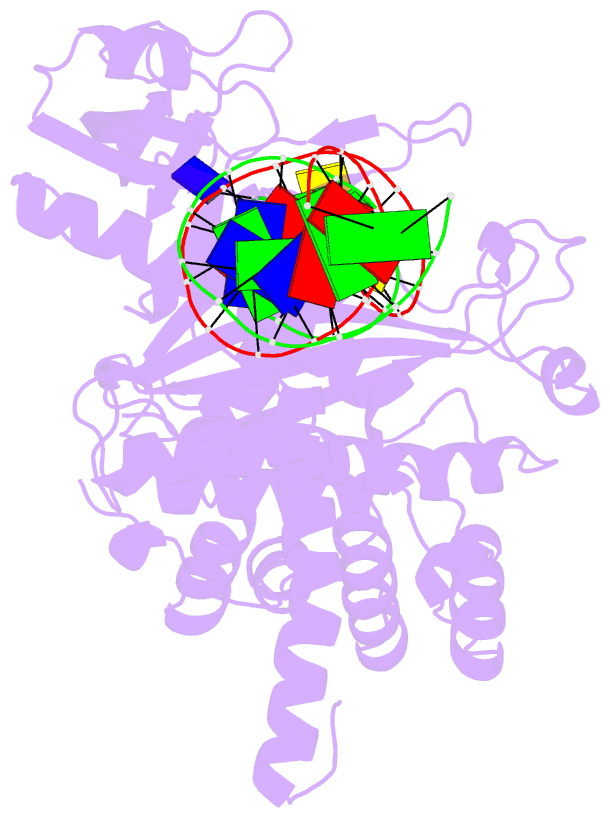
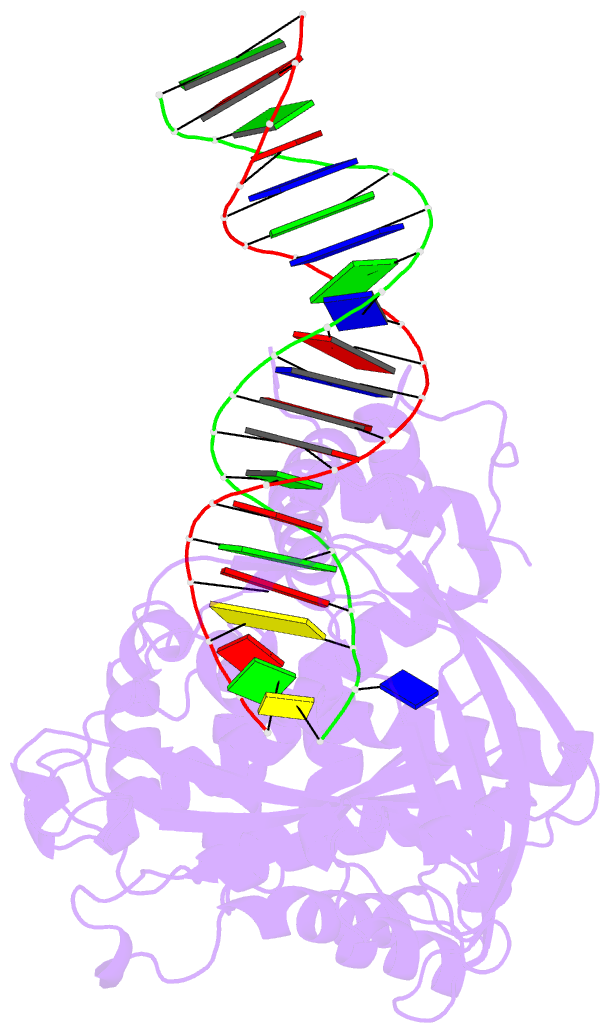
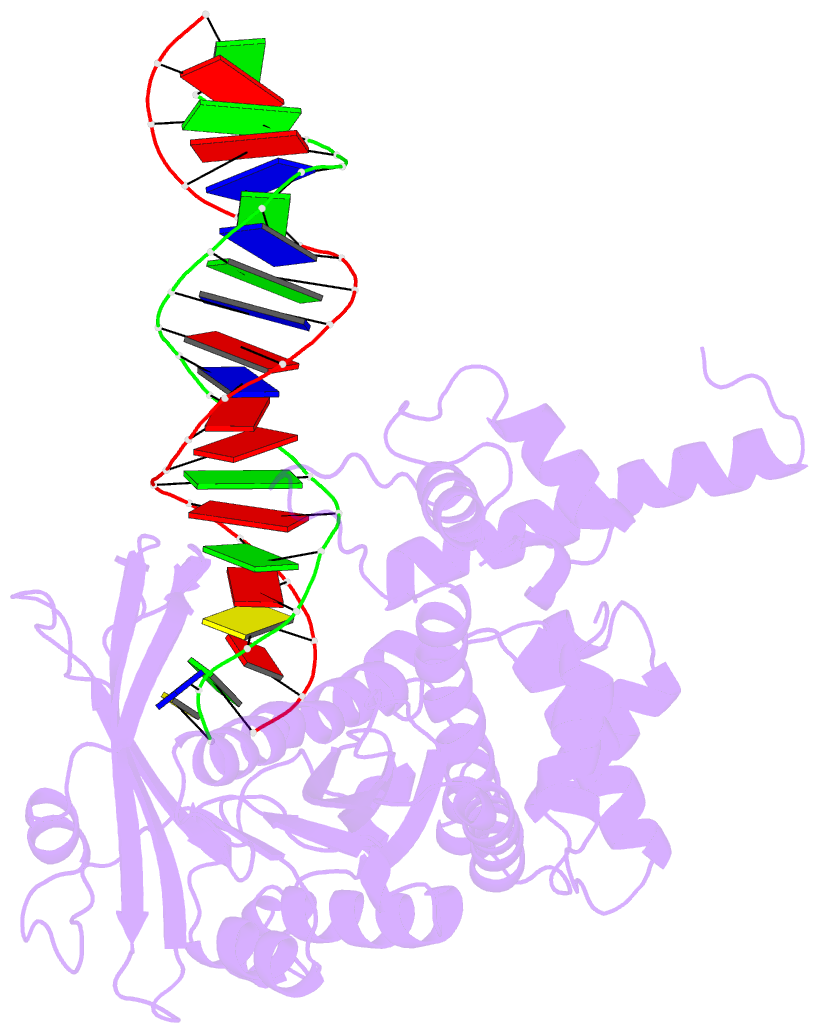
Summary information and primary citation
- PDB-id
- 1mm8; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.8 Å)
- Summary
- Crystal structure of tn5 transposase complexed with me DNA
- Reference
- Steiniger-White M, Bhasin A, Lovell S, Rayment I, Reznikoff WS (2002): "Evidence for "unseen" Transposase--DNA contacts." J.Mol.Biol., 322, 971-982. doi: 10.1016/S0022-2836(02)00877-X.
- Abstract
- In this study, evidence of novel, important interactions between a hyperactive Tn5 transposon recognition end sequence and hyperactive Tn5 transposase (Tnp) are presented. A hyperactive Tn5 end sequence, the mosaic end (ME), was isolated previously. The ME and a wild-type end sequence, the outside end (OE), differ at only three positions, yet transposition on the ME is tenfold higher than on the OE in vivo. Also, transposition on the ME is much more efficient than transposition on the OE in vitro. Here, we show that the decreased activity observed for the OE is caused by a defect in paired ends complex (PEC) formation resulting from the orientation of the A-T base-pair at position 4 of this end. Efficient PEC formation requires an interaction between the C5-methyl group (C5-Me) on the non-transferred strand thymine base at position 4 (T4) and Tnp. PEC formation on nicked substrates is much less affected by the orientation of the A-T base-pair at position 4, indicating that the C5-Me group is important only for steps preceding nicking. A recently determined co-crystal structure of Tn5 Tnp with the ME is discussed and a model explaining possible roles for the base-pair at position 4 is explored.