Summary information and primary citation
- PDB-id
- 1mnb; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- NMR
- Summary
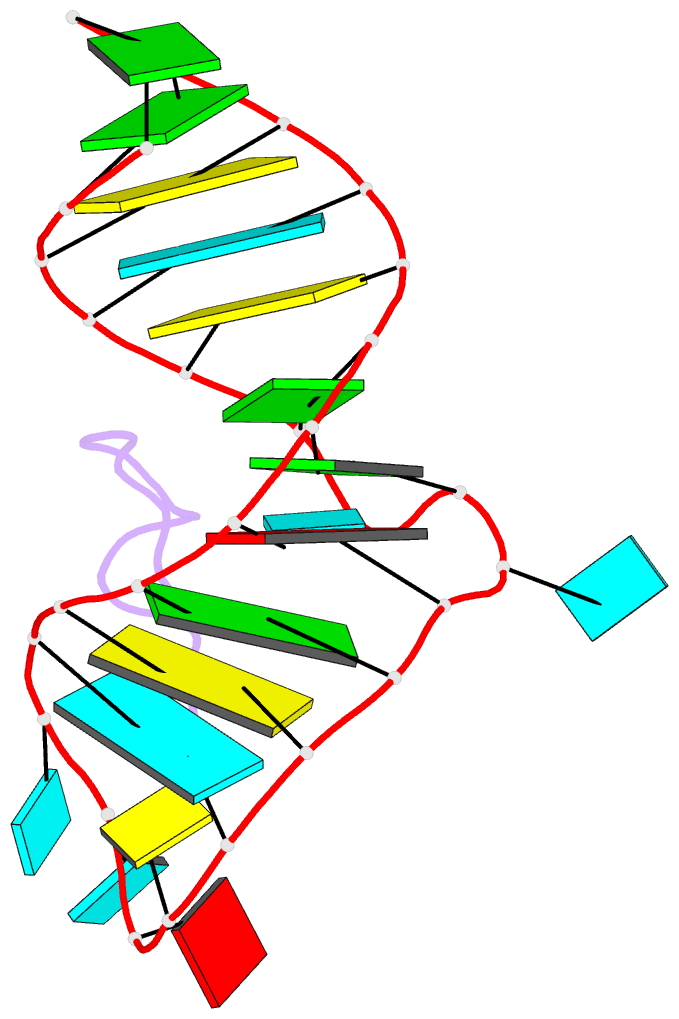
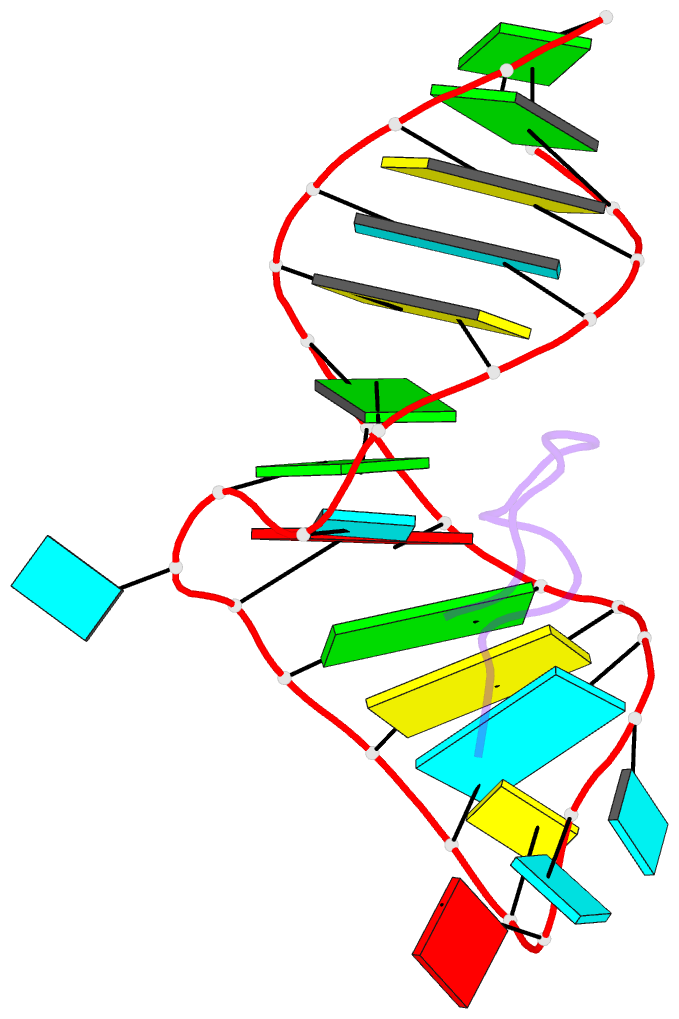
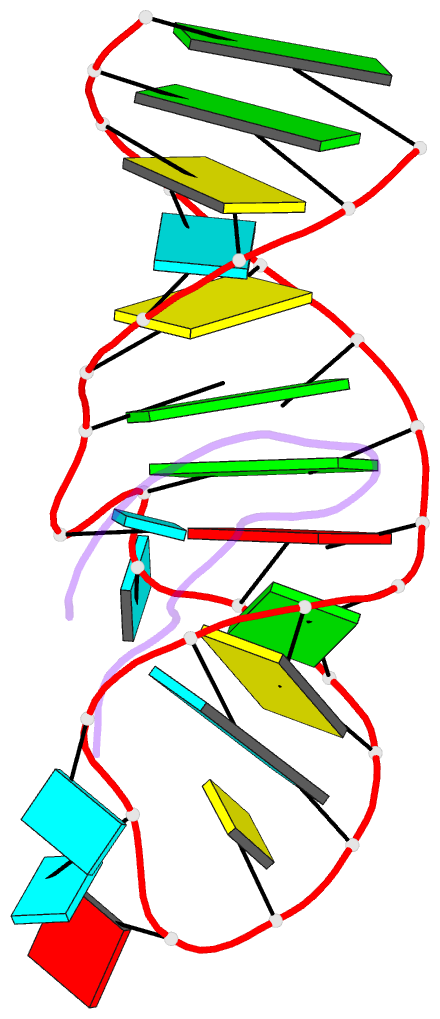
- Biv tat peptide (residues 68-81), NMR, minimized average structure
- Reference
- Puglisi JD, Chen L, Blanchard S, Frankel AD (1995): "Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex." Science, 270, 1200-1203.
- Abstract
- The Tat protein of bovine immunodeficiency virus (BIV) binds to its target RNA, TAR, and activates transcription. A 14-amino acid arginine-rich peptide corresponding to the RNA-binding domain of BIV Tat binds specifically to BIV TAR, and biochemical and in vivo experiments have identified the amino acids and nucleotides required for binding. The solution structure of the RNA-peptide complex has now been determined by nuclear magnetic resonance spectroscopy. TAR forms a virtually continuous A-form helix with two unstacked bulged nucleotides. The peptide adopts a beta-turn conformation and sits in the major groove of the RNA. Specific contacts are apparent between critical amino acids in the peptide and bases and phosphates in the RNA. The structure is consistent with all biochemical data and demonstrates ways in which proteins can recognize the major groove of RNA.