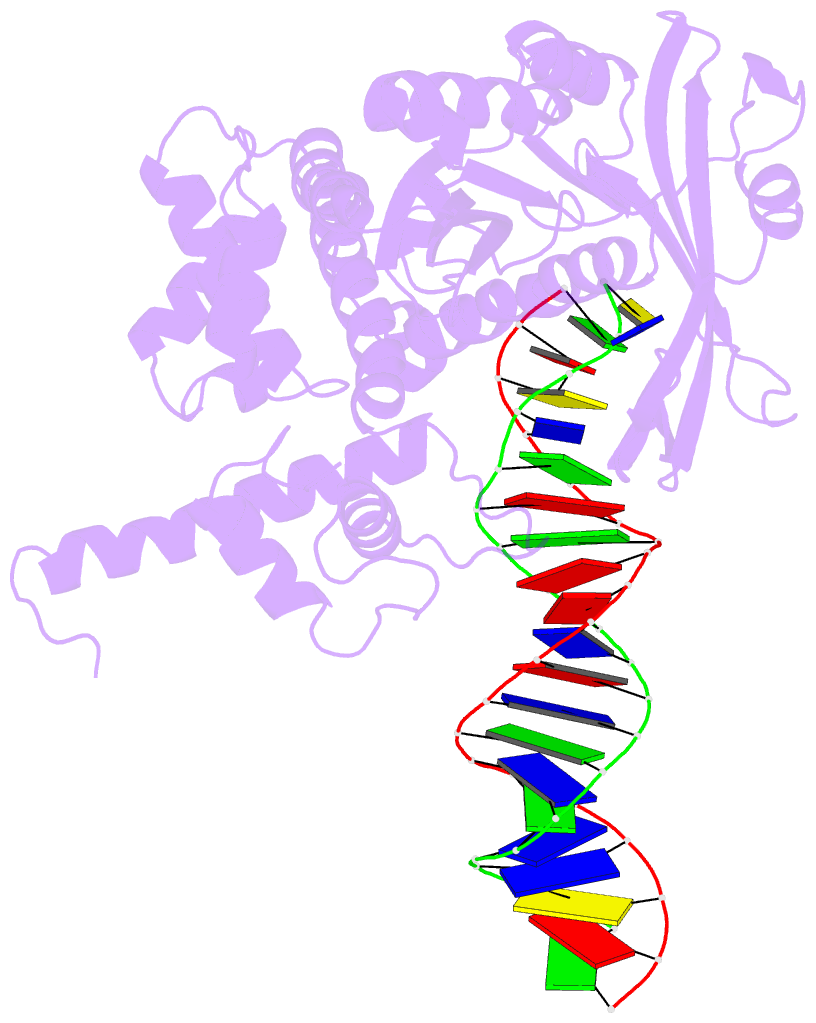
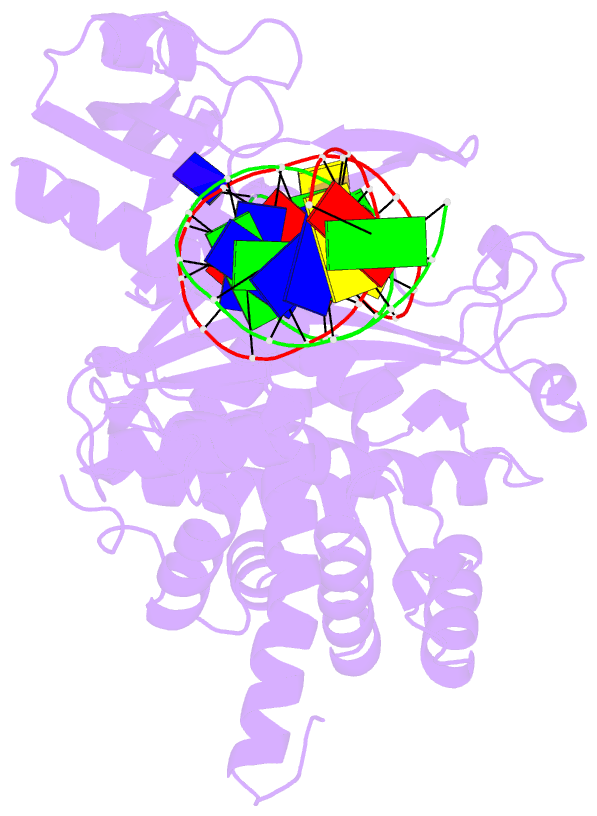
Summary information and primary citation
- PDB-id
- 1muh; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.3 Å)
- Summary
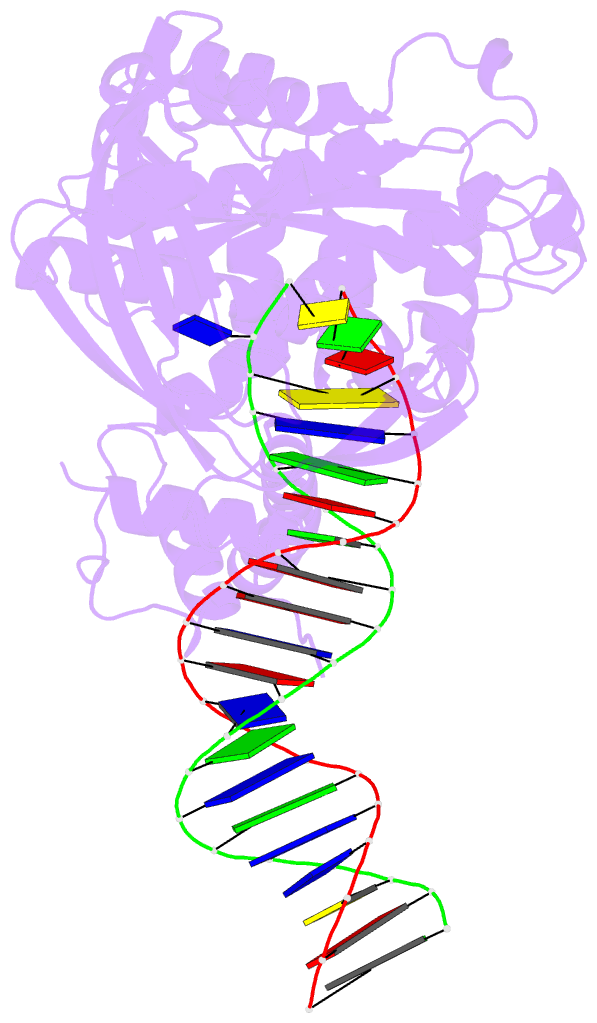
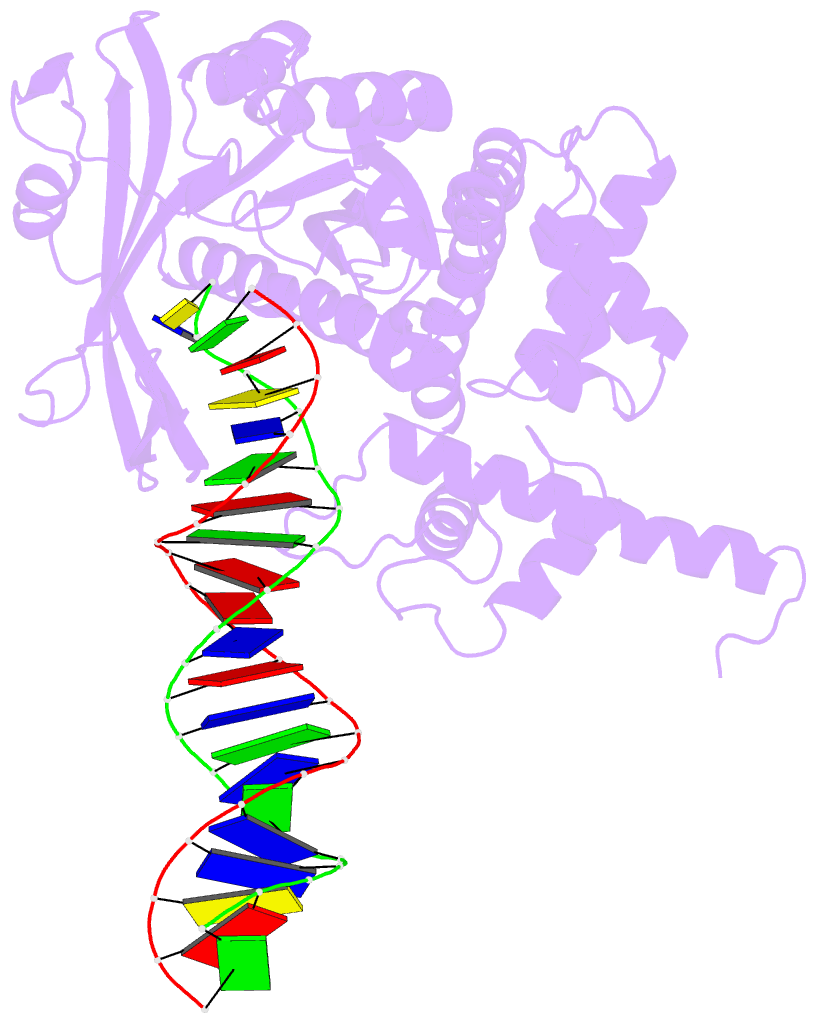
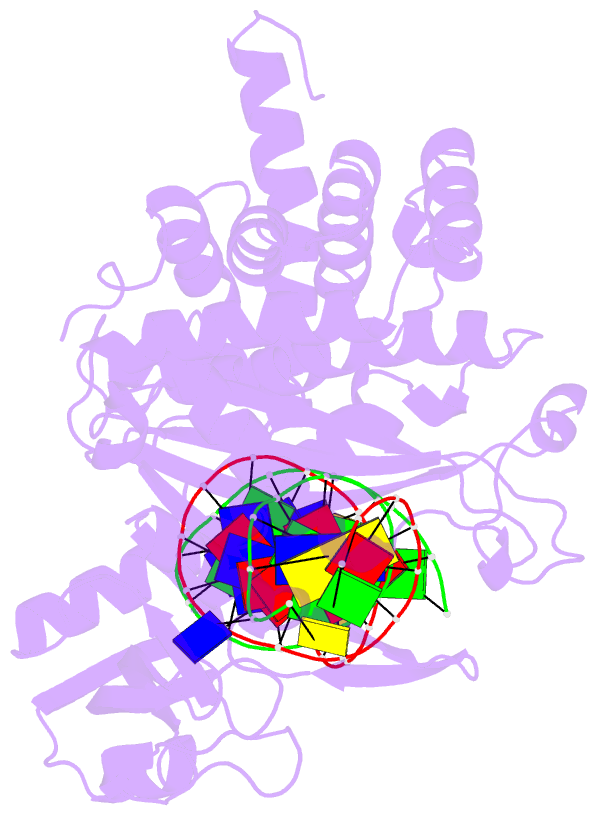
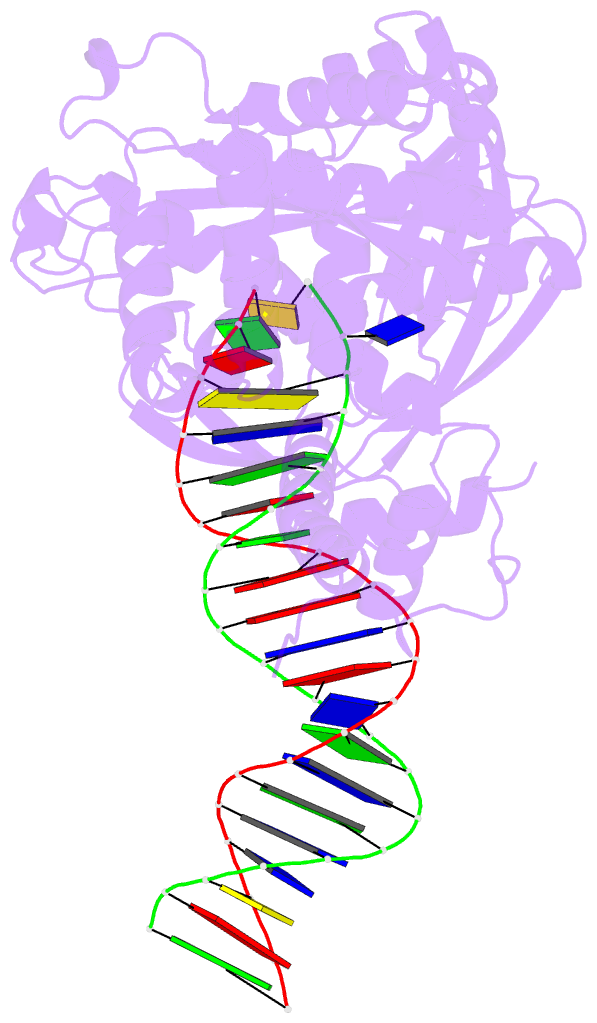
- Crystal structure of tn5 transposase complexed with transposon end DNA
- Reference
- Davies DR, Goryshin IY, Reznikoff WS, Rayment I (2000): "Three-dimensional structure of the Tn5 synaptic complex transposition intermediate." Science, 289, 77-85. doi: 10.1126/science.289.5476.77.
- Abstract
- Genomic evolution has been profoundly influenced by DNA transposition, a process whereby defined DNA segments move freely about the genome. Transposition is mediated by transposases, and similar events are catalyzed by retroviral integrases such as human immunodeficiency virus-1 (HIV-1) integrase. Understanding how these proteins interact with DNA is central to understanding the molecular basis of transposition. We report the three-dimensional structure of prokaryotic Tn5 transposase complexed with Tn5 transposon end DNA determined to 2.3 angstrom resolution. The molecular assembly is dimeric, where each double-stranded DNA molecule is bound by both protein subunits, orienting the transposon ends into the active sites. This structure provides a molecular framework for understanding many aspects of transposition, including the binding of transposon end DNA by one subunit and cleavage by a second, cleavage of two strands of DNA by a single active site via a hairpin intermediate, and strand transfer into target DNA.