Summary information and primary citation
- PDB-id
- 1mw8; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- isomerase
- Method
- X-ray (1.9 Å)
- Summary
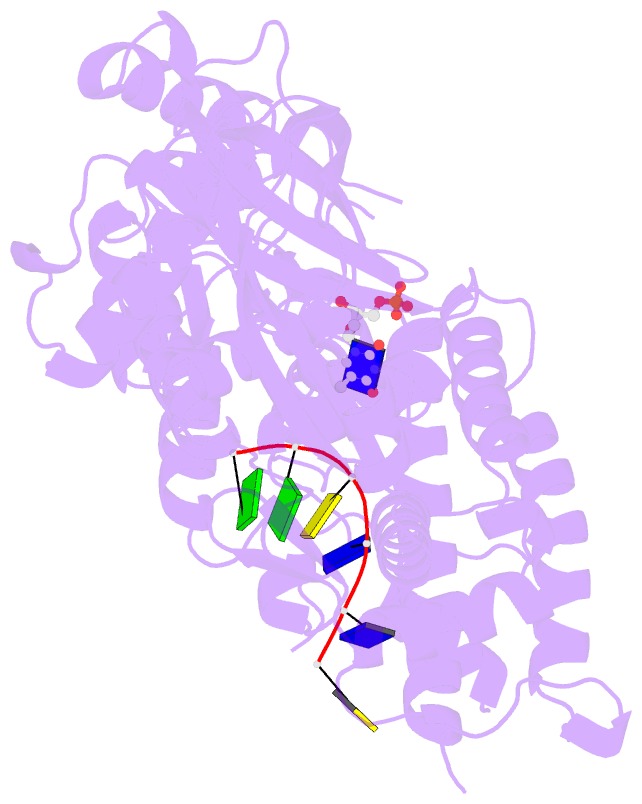
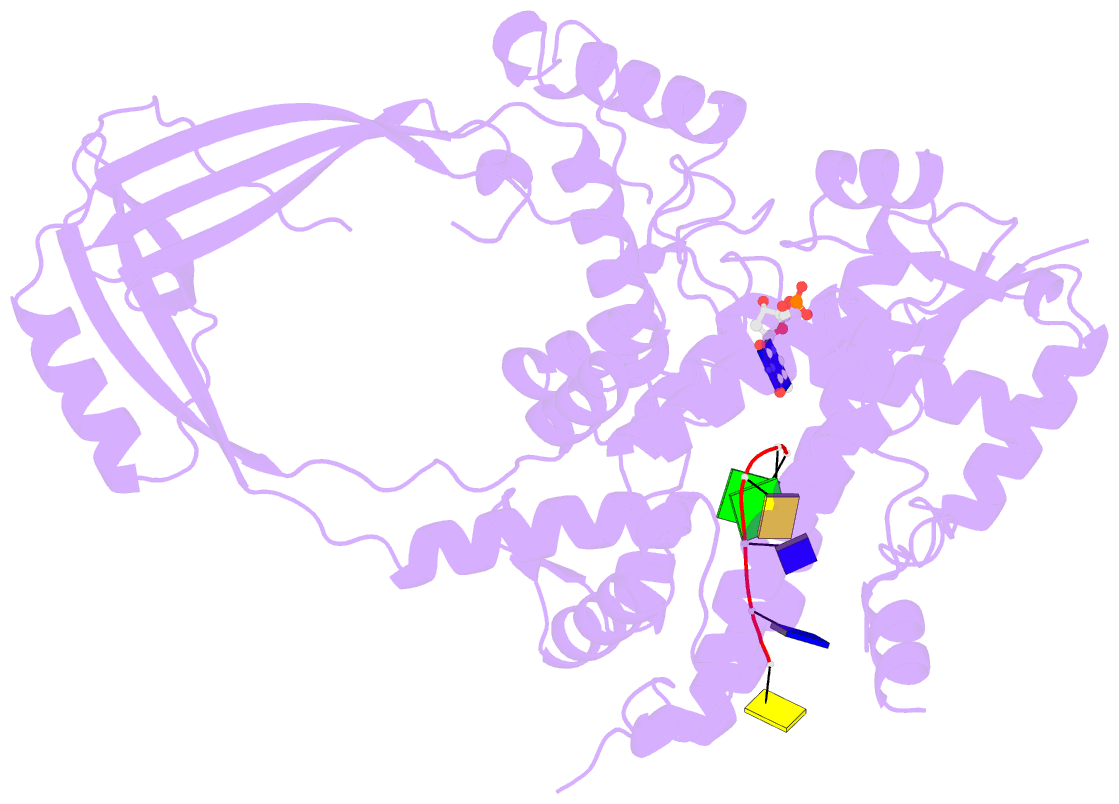
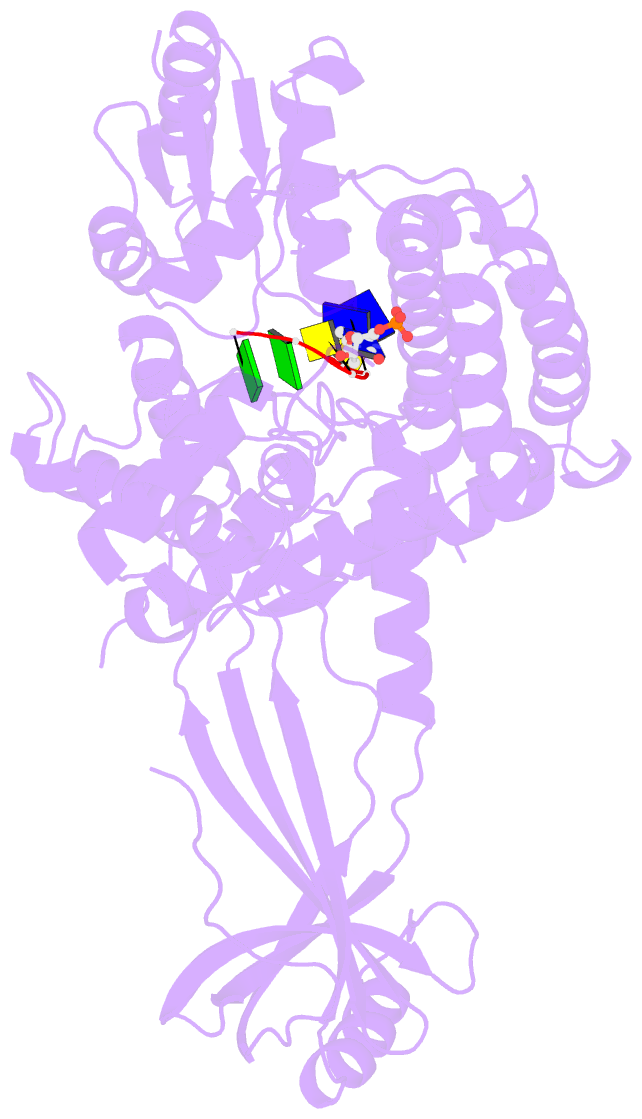
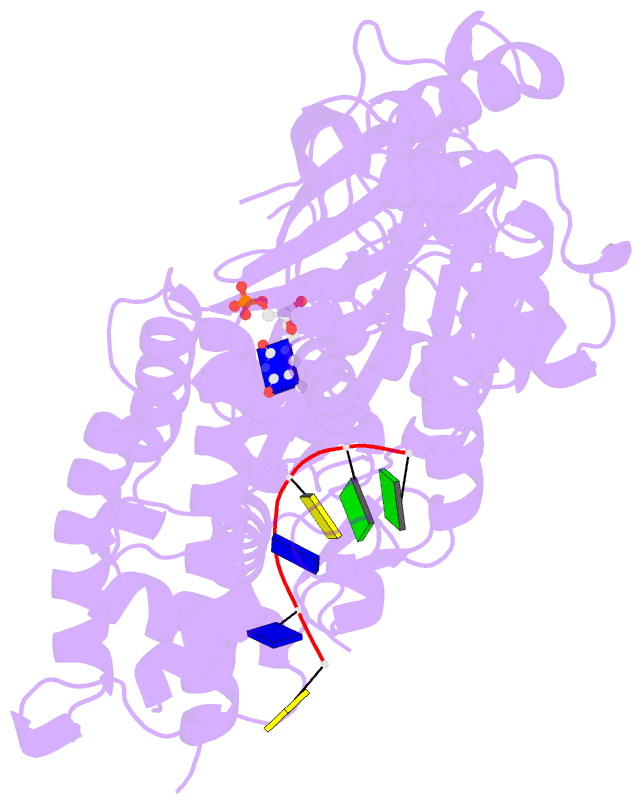
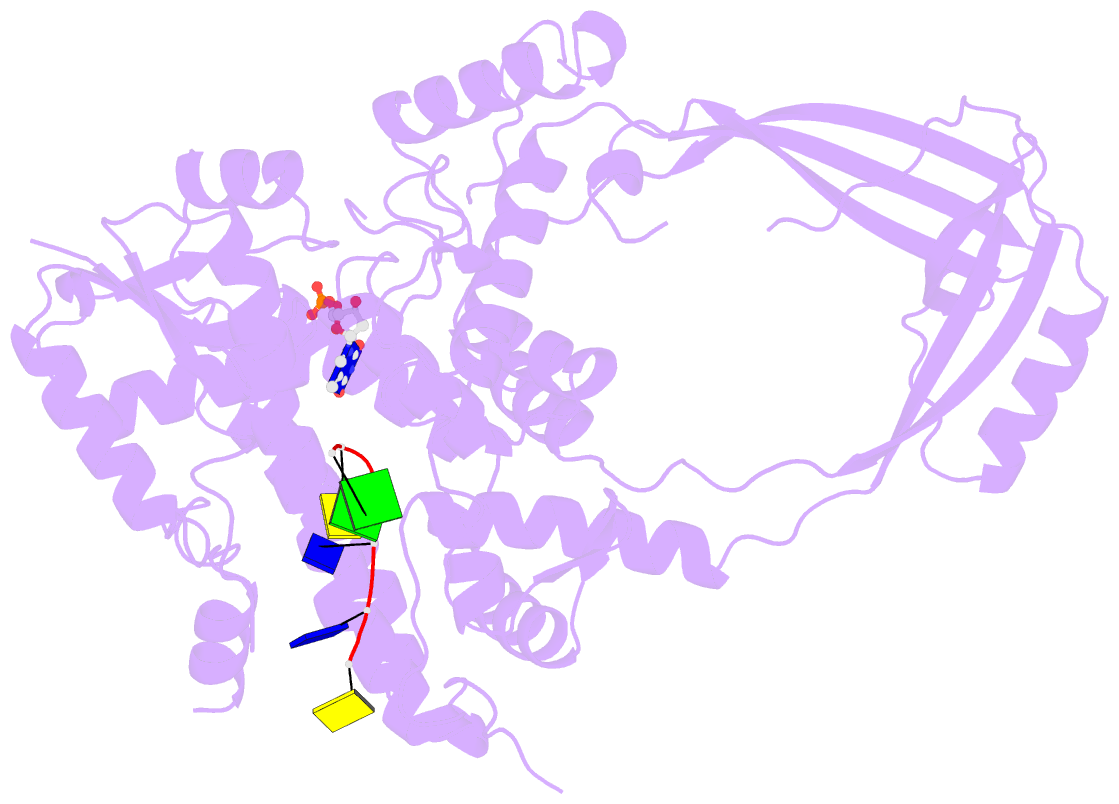
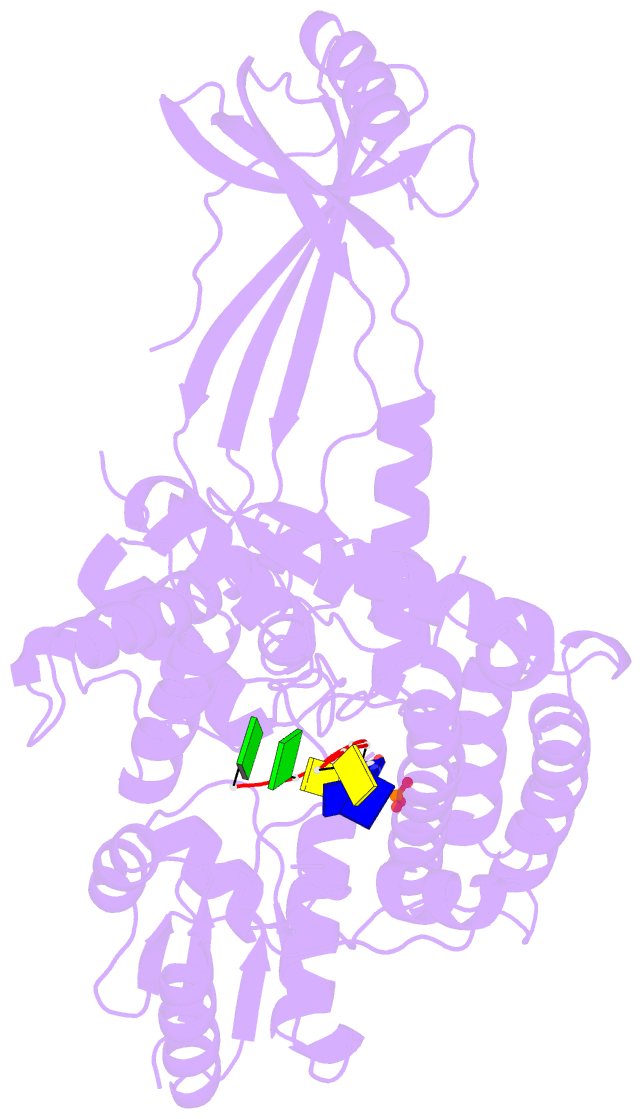
- Crystal structure of a complex between h365r mutant of 67 kda n-terminal fragment of e. coli DNA topoisomerase i and 5'-acttcgggatg-3'
- Reference
- Perry K, Mondragon A (2003): "Structure of a Complex between E. coli DNA Topoisomerase I and Single-Stranded DNA." Structure, 11, 1349-1358. doi: 10.1016/j.str.2003.09.013.
- Abstract
- In order to gain insights into the mechaism of ssDNA binding and recognition by Escherichia coli DNA topoisomerase I, the structure of the 67 kDa N-terminal fragment of topoisomerase I was solved in complex with ssDNA. The structure reveals a new conformational stage in the multistep catalytic cycle of type IA topoisomerases. In the structure, the ssDNA binding groove leading to the active site is occupied, but the active site is not fully formed. Large conformational changes are not seen; instead, a single helix parallel to the ssDNA binding groove shifts to clamp the ssDNA. The structure helps clarify the temporal sequence of conformational events, starting from an initial empty enzyme and proceeding to a ssDNA-occupied and catalytically competent active site.