Summary information and primary citation
- PDB-id
-
1n32;
DSSR-derived features in text and
JSON formats; DNAproDB
- Class
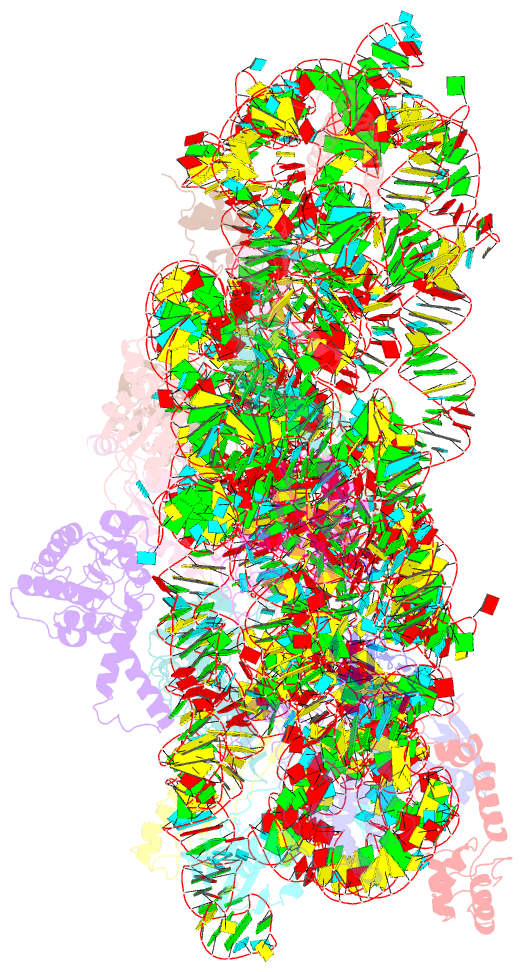
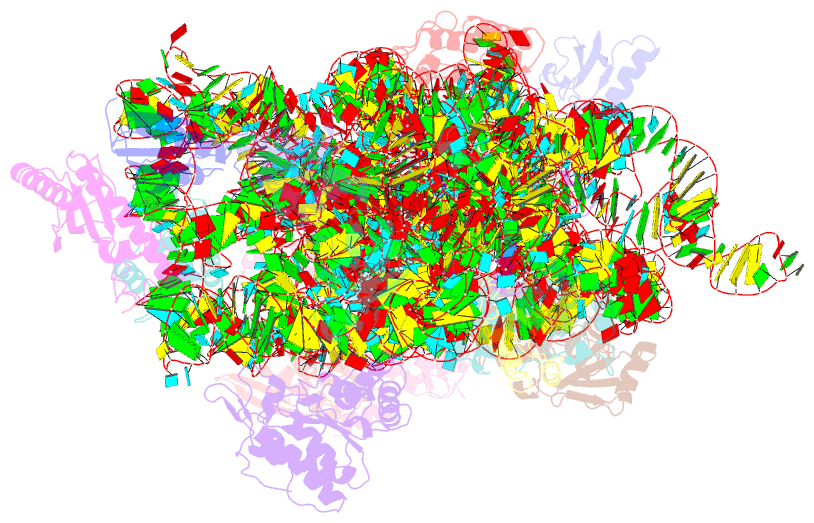
- ribosome
- Method
- X-ray (3.0 Å)
- Summary
- Structure of the thermus thermophilus 30s ribosomal
subunit bound to codon and near-cognate transfer RNA
anticodon stem-loop mismatched at the first codon position
at the a site with paromomycin
- Reference
-
Ogle JM, Murphy IV FV, Tarry MJ, Ramakrishnan V (2002):
"Selection
of tRNA by the Ribosome Requires a Transition from an
Open to a Closed Form."
Cell(Cambridge,Mass.), 111,
721-732. doi: 10.1016/S0092-8674(02)01086-3.
- Abstract
- A structural and mechanistic explanation for the
selection of tRNAs by the ribosome has been elusive. Here,
we report crystal structures of the 30S ribosomal subunit
with codon and near-cognate tRNA anticodon stem loops bound
at the decoding center and compare affinities of equivalent
complexes in solution. In ribosomal interactions with
near-cognate tRNA, deviation from Watson-Crick geometry
results in uncompensated desolvation of hydrogen-bonding
partners at the codon-anticodon minor groove. As a result,
the transition to a closed form of the 30S induced by
cognate tRNA is unfavorable for near-cognate tRNA unless
paromomycin induces part of the rearrangement. We conclude
that stabilization of a closed 30S conformation is required
for tRNA selection, and thereby structurally rationalize
much previous data on translational fidelity.