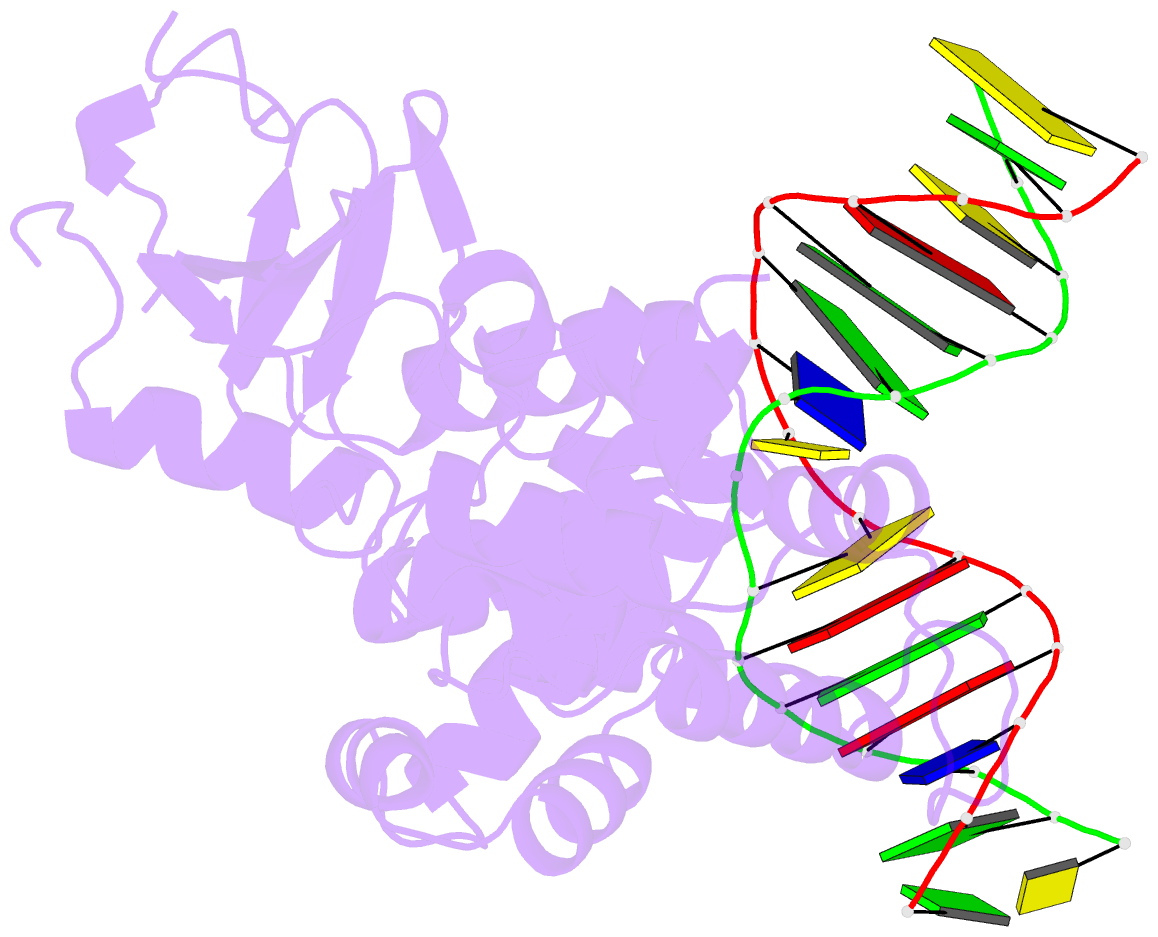
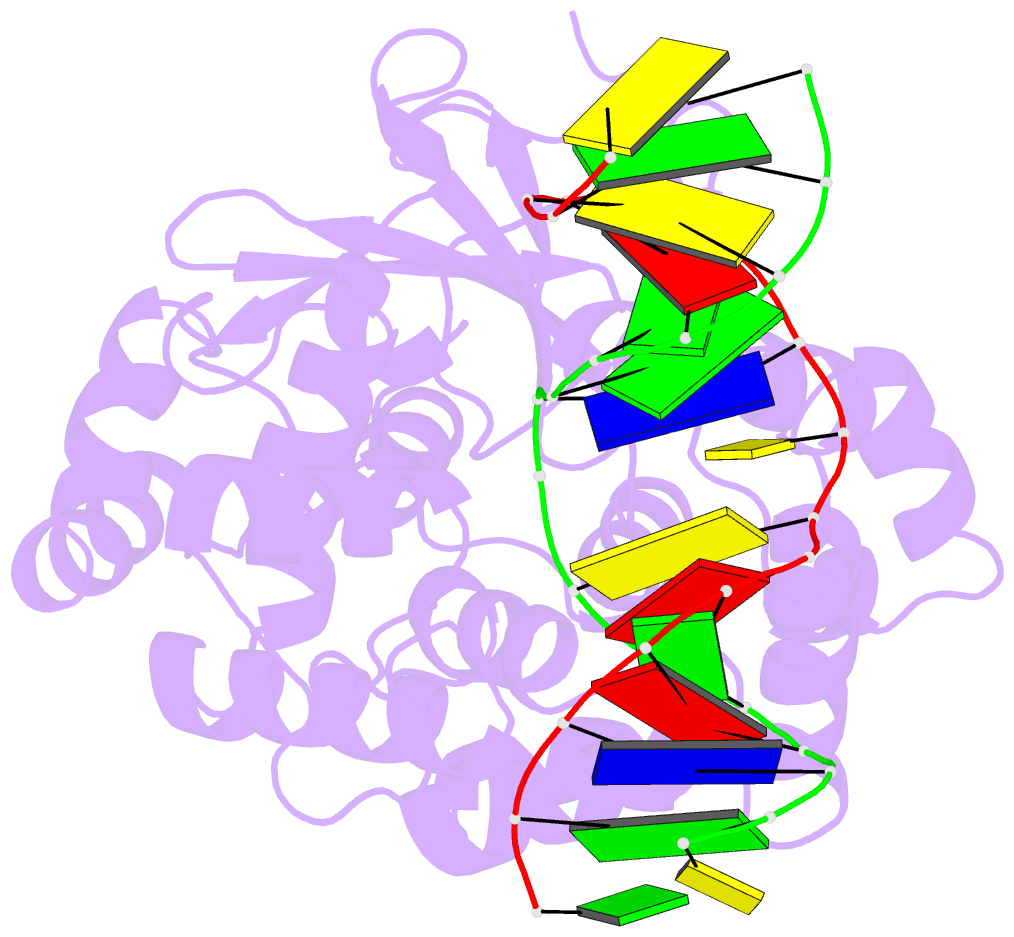
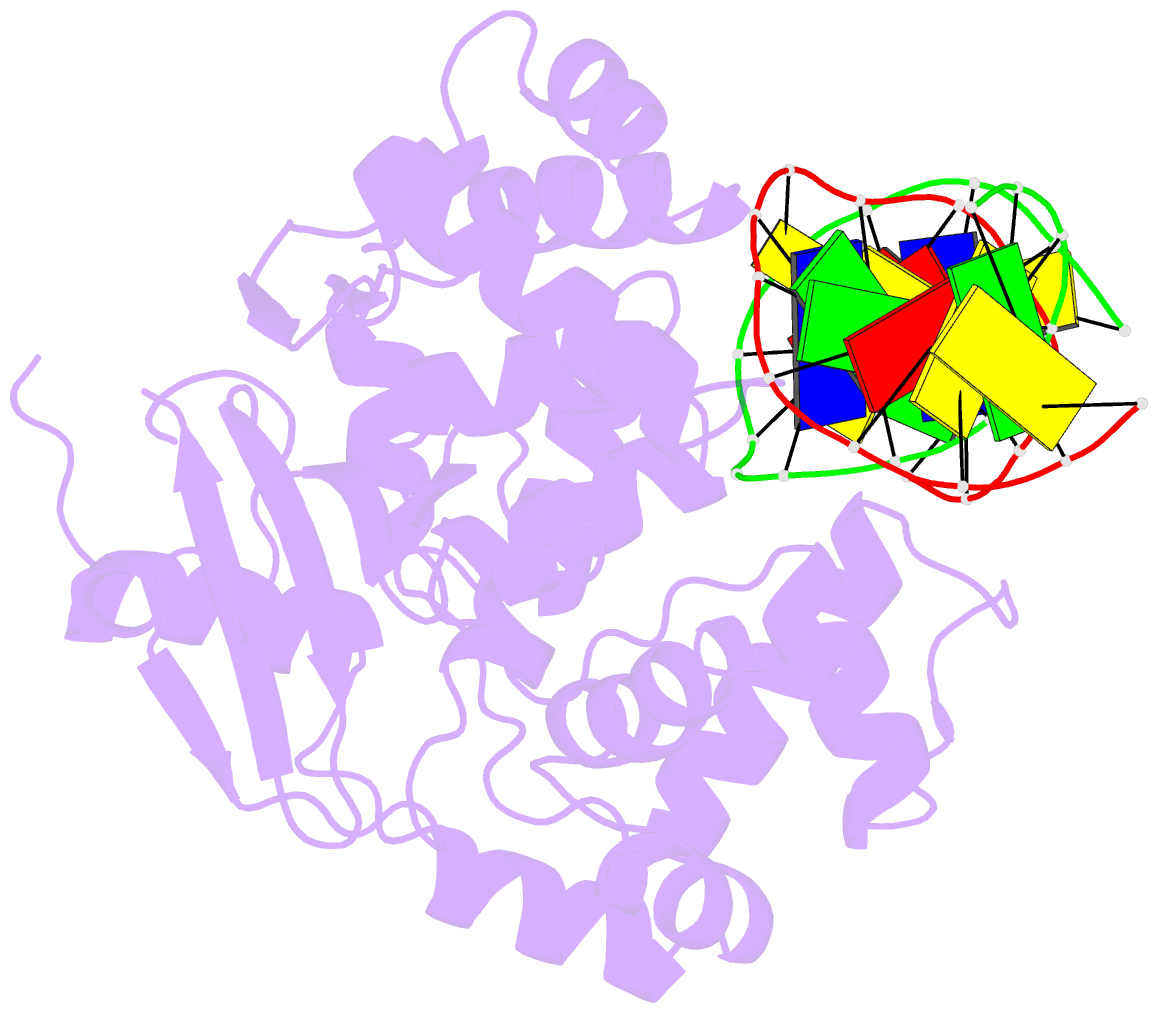
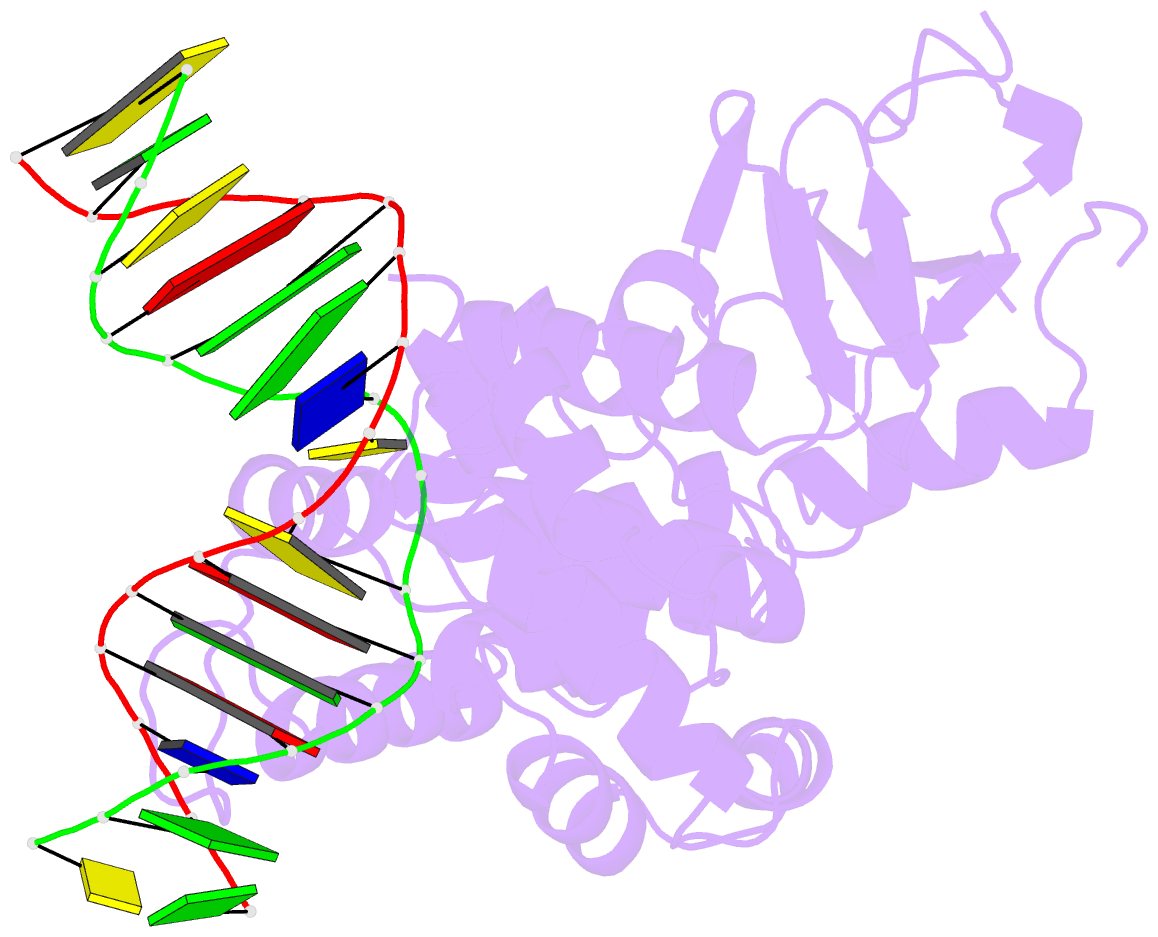
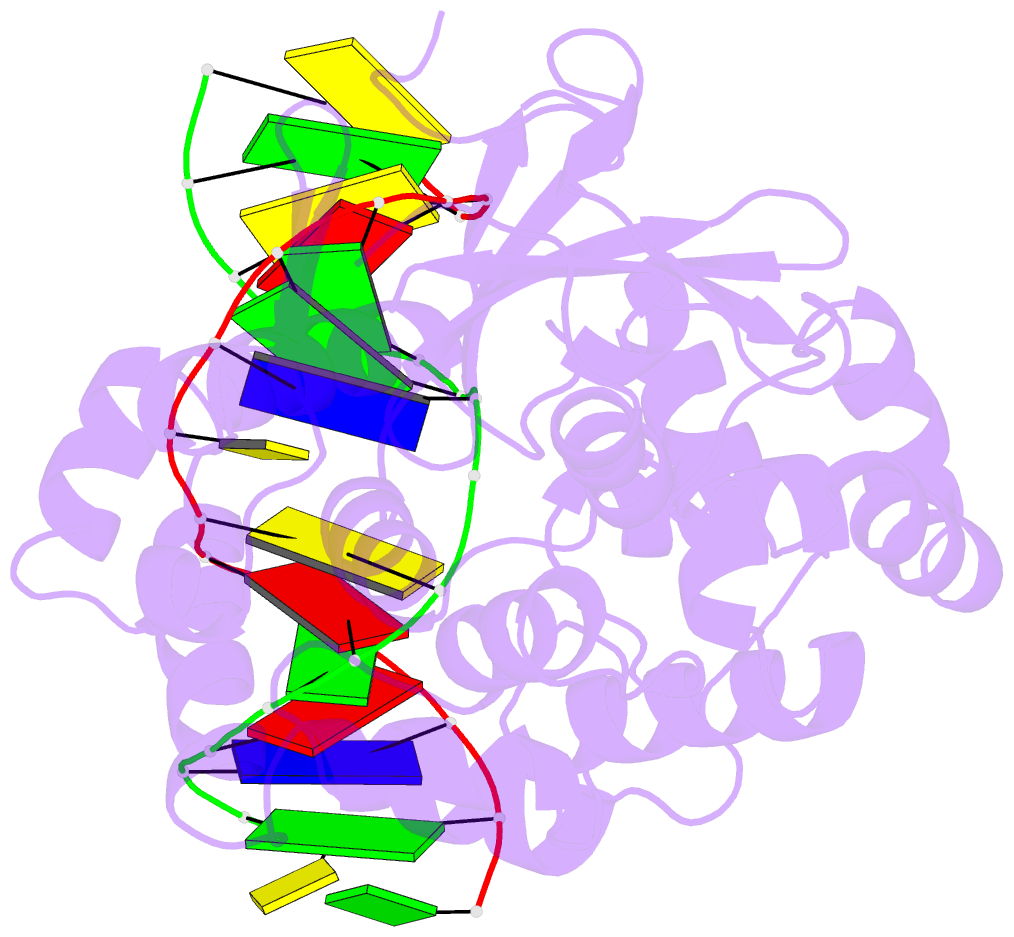
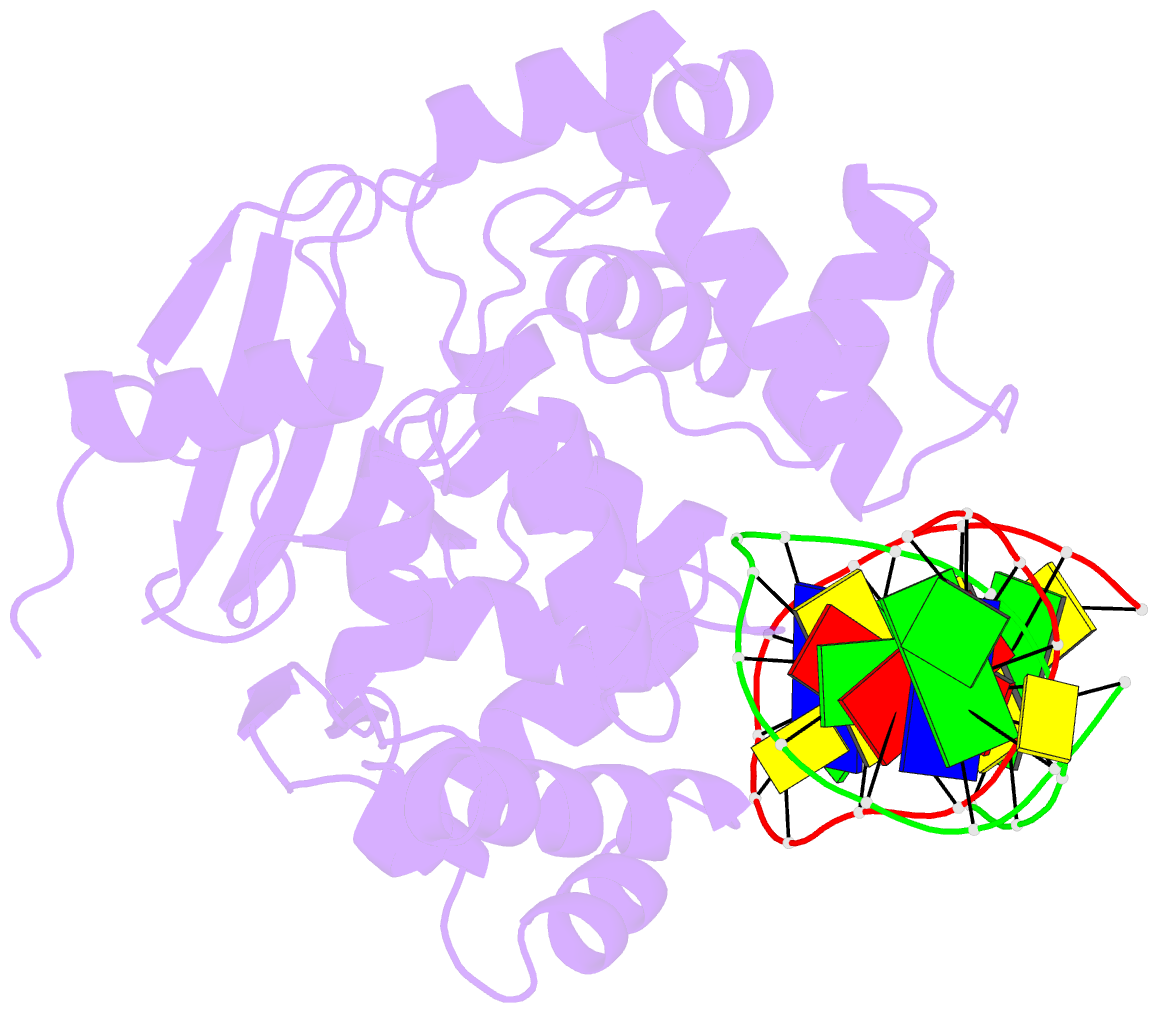
Summary information and primary citation
- PDB-id
- 1n39; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase, lyase-DNA
- Method
- X-ray (2.2 Å)
- Summary
- Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase
- Reference
- Norman DP, Chung SJ, Verdine GL (2003): "Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase." Biochemistry, 42, 1564-1572. doi: 10.1021/bi026823d.
- Abstract
- Members of the HhH-GPD superfamily of DNA glycosylases are responsible for the recognition and removal of damaged nucleobases from DNA. The hallmark of these proteins is a motif comprising a helix-hairpin-helix followed by a Gly/Pro-rich loop and terminating in an invariant, catalytically essential aspartic acid residue. In this study, we have probed the role of this Asp in human 8-oxoguanine DNA glycosylase (hOgg1) by mutating it to Asn (D268N), Glu (D268E), and Gln (D268Q). We show that this aspartate plays a dual role, acting both as an N-terminal alpha-helix cap and as a critical residue for catalysis of both base excision and DNA strand cleavage by hOgg1. Mutation of this residue to asparagine, another helix-capping residue, preserves stability of the protein while drastically reducing enzymatic activity. A crystal structure of this mutant is the first to reveal the active site nucleophile Lys249 in the presence of lesion-containing DNA; this structure offers a tantalizing suggestion that base excision may occur by cleavage of the glycosidic bond and then attachment of Lys249. Mutation of the aspartic acid to glutamine and glutamic acid destabilizes the protein fold to a significant extent but, surprisingly, preserves catalytic activity. Crystal structures of these mutants complexed with an unreactive abasic site in DNA reveal these residues to adopt a sterically disfavored helix-capping conformation.