Summary information and primary citation
- PDB-id
- 1n3e; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.5 Å)
- Summary
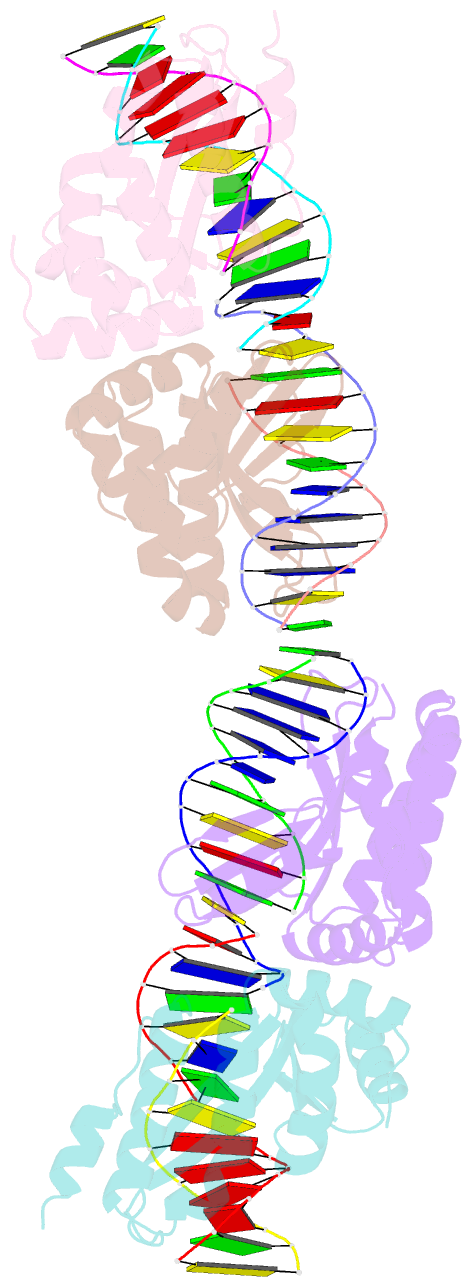
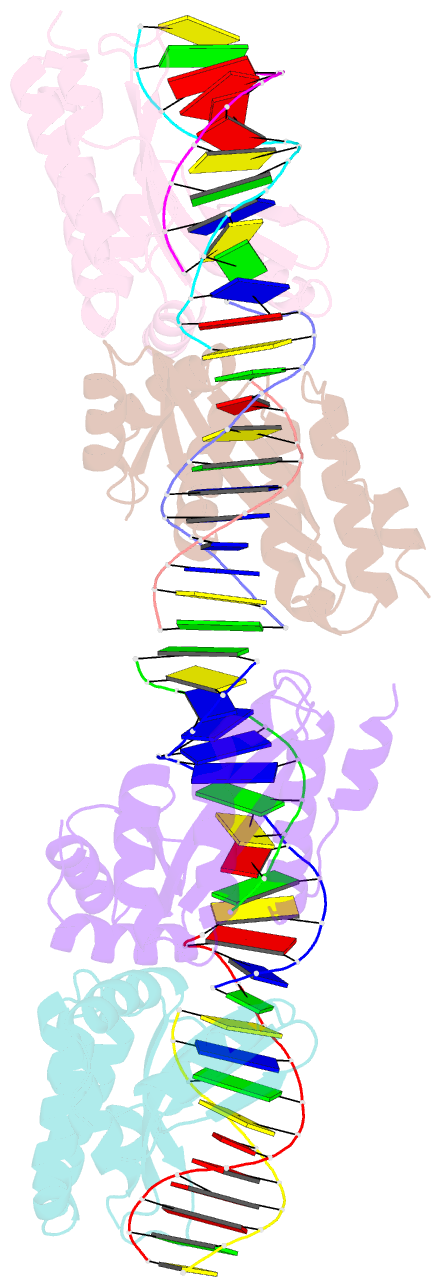
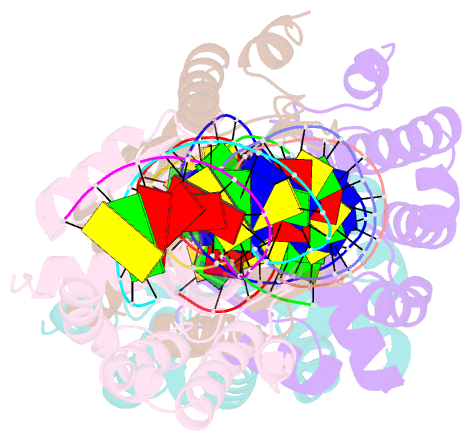
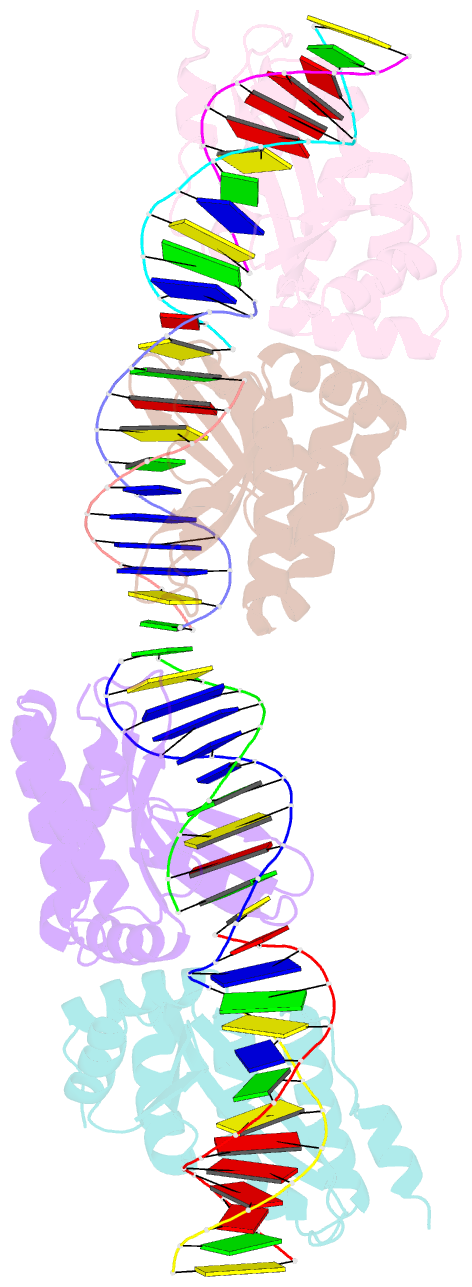
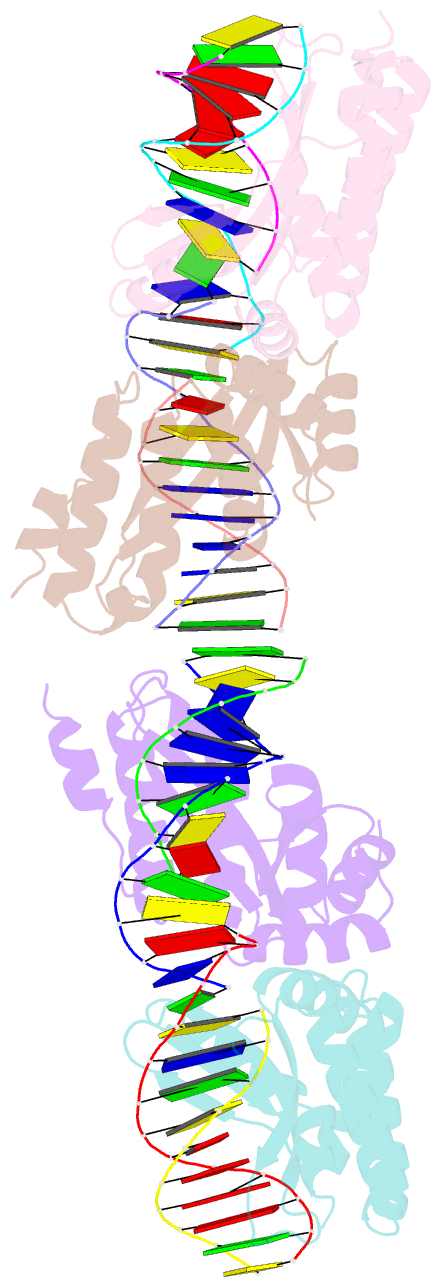
- Crystal structure of i-crei bound to a palindromic DNA sequence i (palindrome of left side of wildtype DNA target sequence)
- Reference
- Chevalier B, Turmel M, Lemieux C, Monnat RJ, Stoddard BL (2003): "Flexible DNA Target Site Recognition by Divergent Homing Endonuclease Isoschizomers I-CreI and I-MsoI." J.Mol.Biol., 329, 253-269. doi: 10.1016/S0022-2836(03)00447-9.
- Abstract
- Homing endonucleases are highly specific catalysts of DNA strand breaks that induce the transposition of mobile intervening sequences containing the endonuclease open reading frame. These enzymes recognize long DNA targets while tolerating individual sequence polymorphisms within those sites. Sequences of the homing endonucleases themselves diversify to a great extent after founding intron invasion events, generating highly divergent enzymes that recognize similar target sequences. Here, we visualize the mechanism of flexible DNA recognition and the pattern of structural divergence displayed by two homing endonuclease isoschizomers. We determined structures of I-CreI bound to two DNA target sites that differ at eight of 22 base-pairs, and the structure of an isoschizomer, I-MsoI, bound to a nearly identical DNA target site. This study illustrates several principles governing promiscuous base-pair recognition by DNA-binding proteins, and demonstrates that the isoschizomers display strikingly different protein/DNA contacts. The structures allow us to determine the information content at individual positions in the binding site as a function of the distribution of direct and water-mediated contacts to nucleotide bases, and provide an evolutionary snapshot of endonucleases at an early stage of divergence in their target specificity.