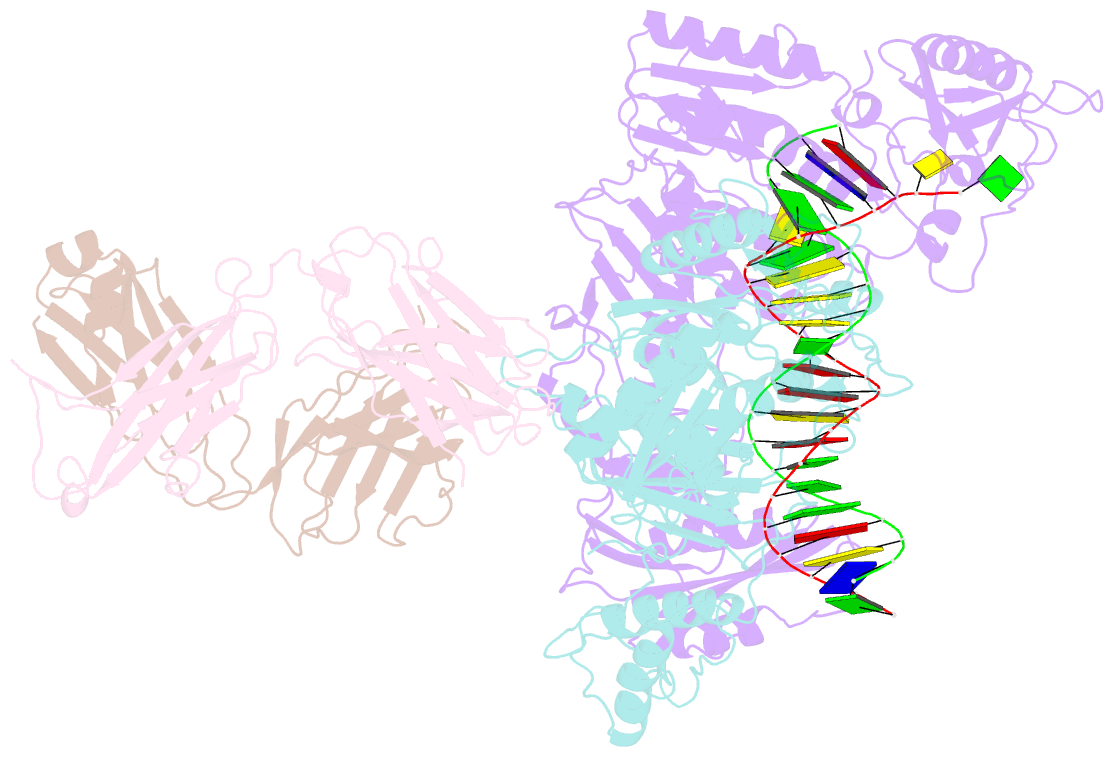
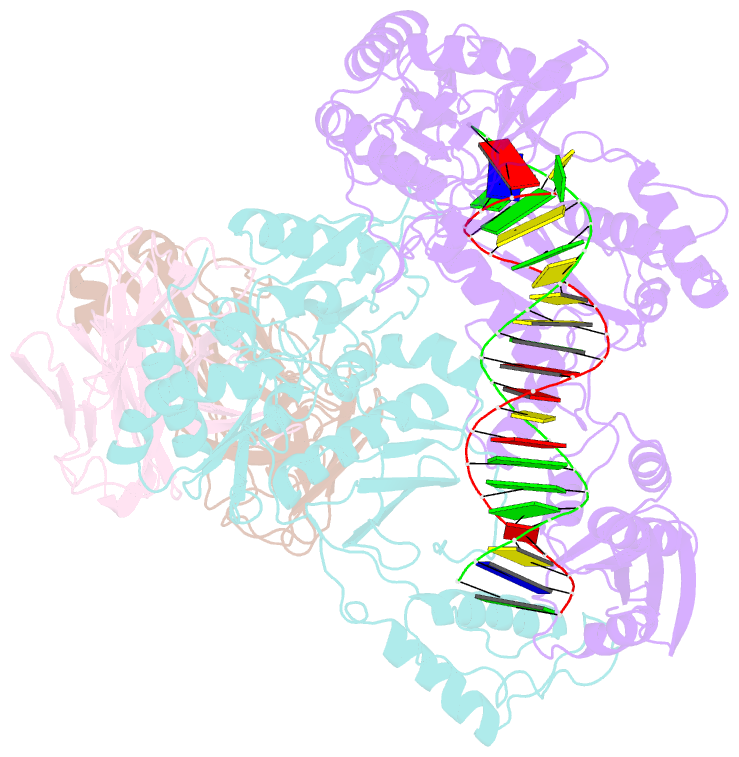
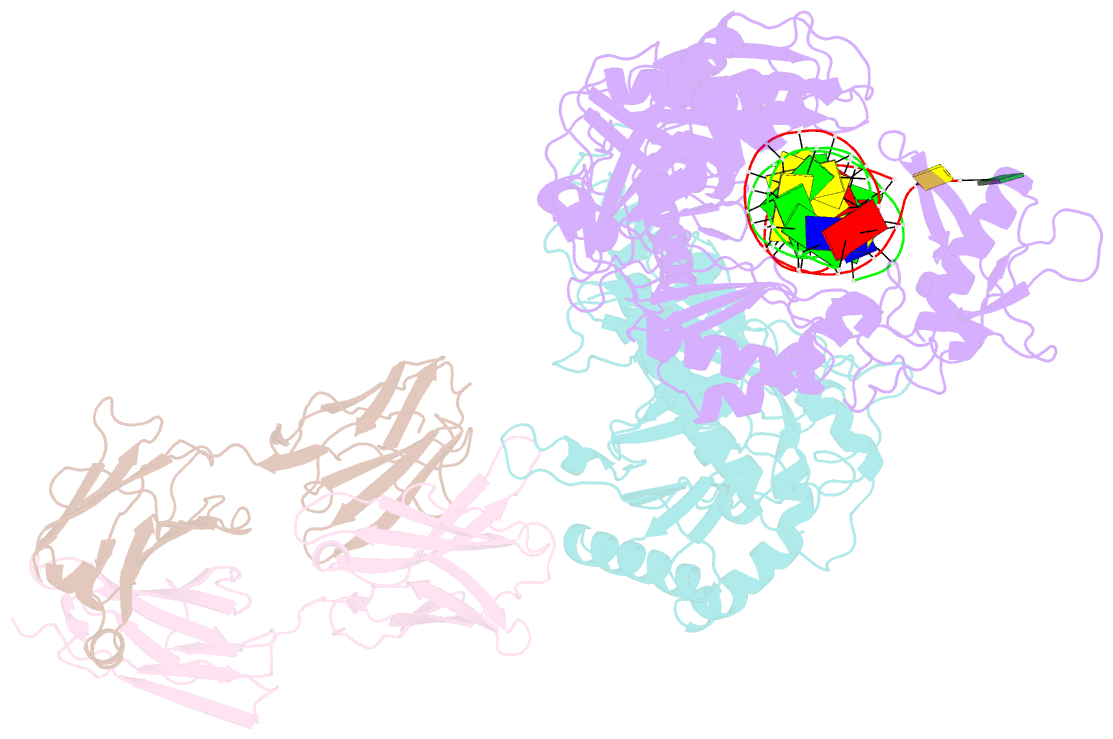
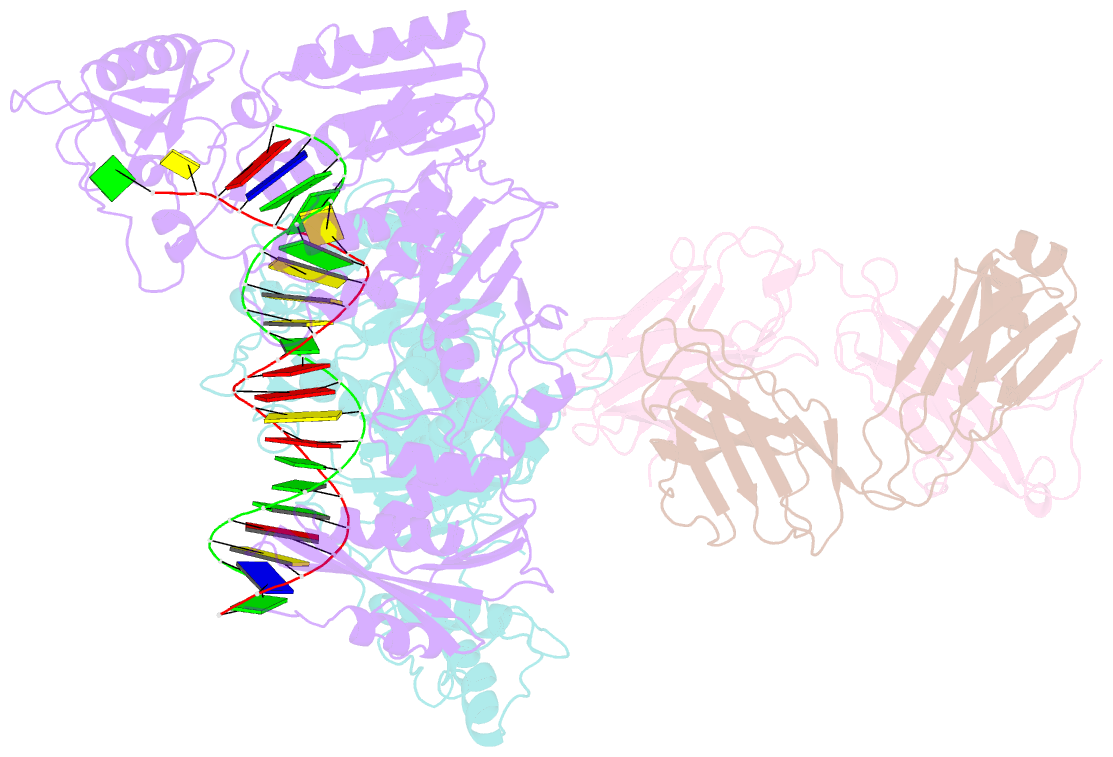
Summary information and primary citation
- PDB-id
- 1n6q; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-immune system-DNA
- Method
- X-ray (3.0 Å)
- Summary
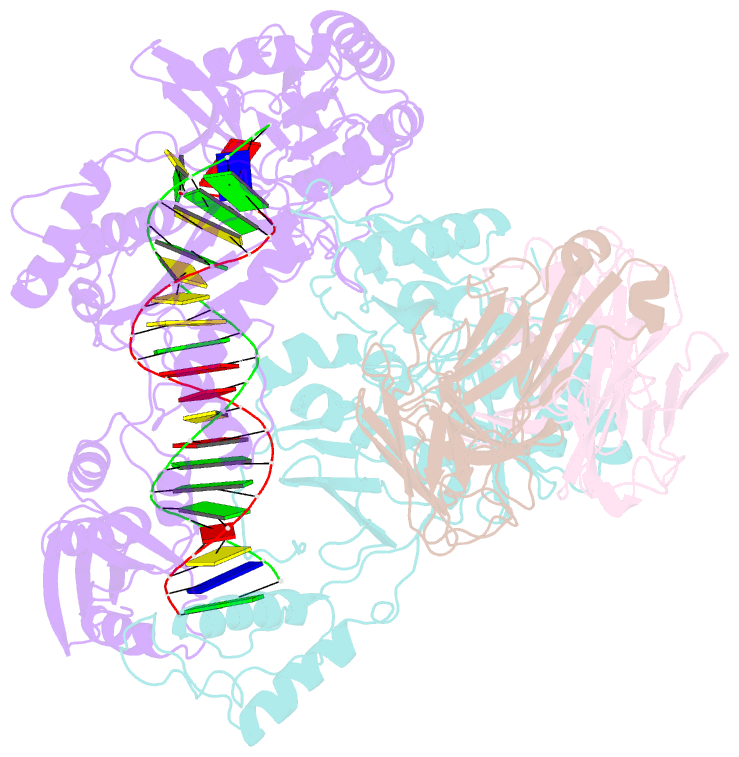
- Hiv-1 reverse transcriptase crosslinked to pre-translocation aztmp-terminated DNA (complex n)
- Reference
- Sarafianos SG, Clark Jr AD, Das K, Tuske S, Birktoft JJ, Ilankumaran I, Ramesha AR, Sayer JM, Jerina DM, Boyer PL, Hughes SH, Arnold E (2002): "Structures of HIV-1 Reverse Transcriptase with Pre- and Post-translocation AZTMP-terminated DNA." Embo J., 21, 6614-6624. doi: 10.1093/emboj/cdf637.
- Abstract
- AZT (3'-azido-3'-deoxythymidine) resistance involves the enhanced excision of AZTMP from the end of the primer strand by HIV-1 reverse transcriptase. This reaction can occur when an AZTMP-terminated primer is bound at the nucleotide-binding site (pre-translocation complex N) but not at the 'priming' site (post-translocation complex P). We determined the crystal structures of N and P complexes at 3.0 and 3.1 A resolution. These structures provide insight into the structural basis of AZTMP excision and the mechanism of translocation. Docking of a dNTP in the P complex structure suggests steric crowding in forming a stable ternary complex that should increase the relative amount of the N complex, which is the substrate for excision. Structural differences between complexes N and P suggest that the conserved YMDD loop is involved in translocation, acting as a springboard that helps to propel the primer terminus from the N to the P site after dNMP incorporation.