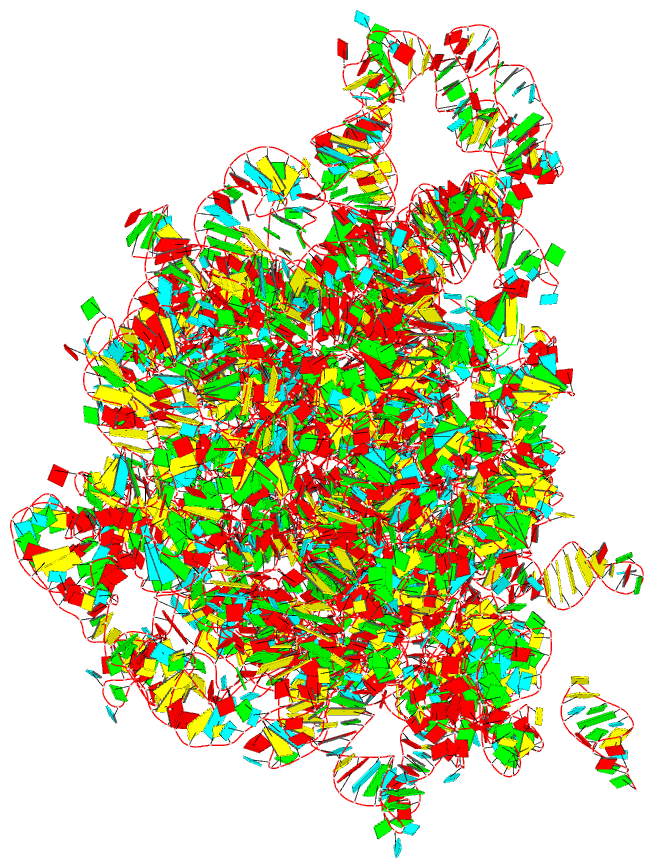
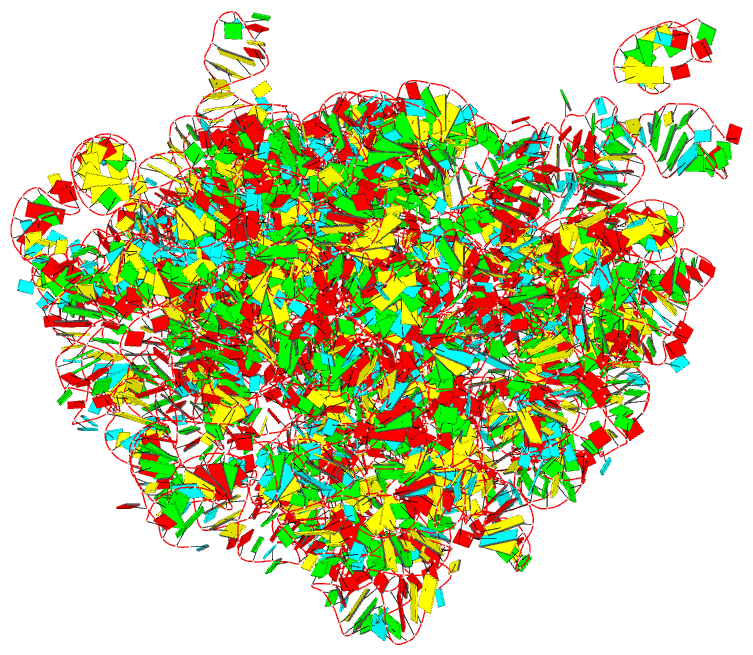
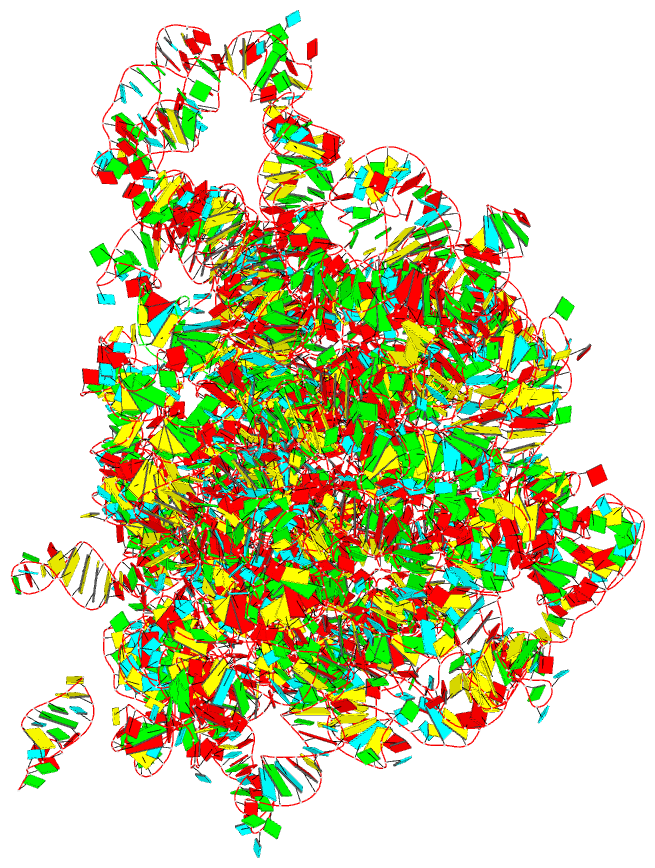
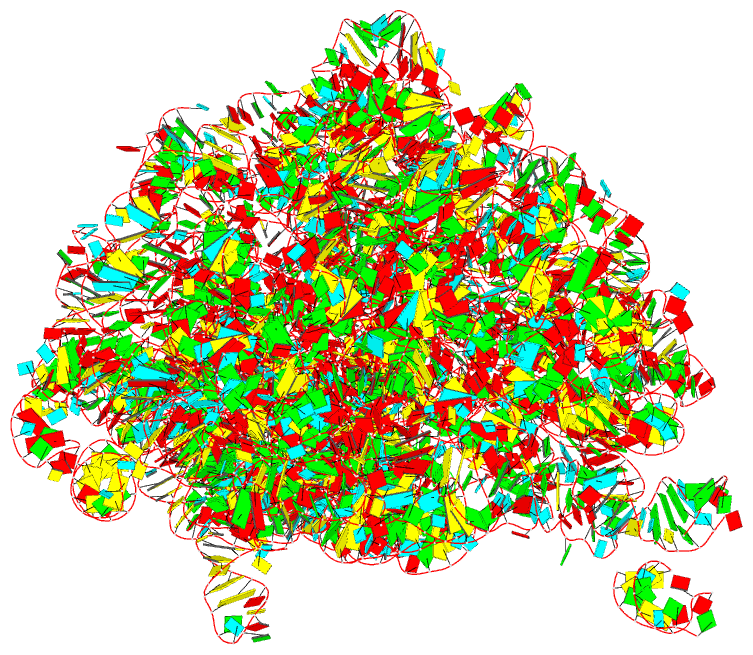
Summary information and primary citation
- PDB-id
- 1njp; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- X-ray (3.5 Å)
- Summary
- The crystal structure of the 50s large ribosomal subunit from deinococcus radiodurans complexed with a trna acceptor stem mimic (asm)
- Reference
- Bashan A, Agmon I, Zarivatch R, Schluenzen F, Harms JM, Berisio R, Bartels H, Franceschi F, Auerbach T, Hansen HA, Kossoy E, Kessler M, Yonath A (2003): "Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression." Mol.Cell, 11, 91-102. doi: 10.1016/S1097-2765(03)00009-1.
- Abstract
- Crystal structures of tRNA mimics complexed with the large ribosomal subunit of Deinococcus radiodurans indicate that remote interactions determine the precise orientation of tRNA in the peptidyl-transferase center (PTC). The PTC tolerates various orientations of puromycin derivatives and its flexibility allows the conformational rearrangements required for peptide-bond formation. Sparsomycin binds to A2602 and alters the PTC conformation. H69, the intersubunit-bridge connecting the PTC and decoding site, may also participate in tRNA placement and translocation. A spiral rotation of the 3' end of the A-site tRNA around a 2-fold axis of symmetry identified within the PTC suggests a unified ribosomal machinery for peptide-bond formation, A-to-P-site translocation, and entrance of nascent proteins into the exit tunnel. Similar 2-fold related regions, detected in all known structures of large ribosomal subunits, indicate the universality of this mechanism.