Summary information and primary citation
- PDB-id
- 1o3s; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- gene regulation-DNA
- Method
- X-ray (3.0 Å)
- Summary
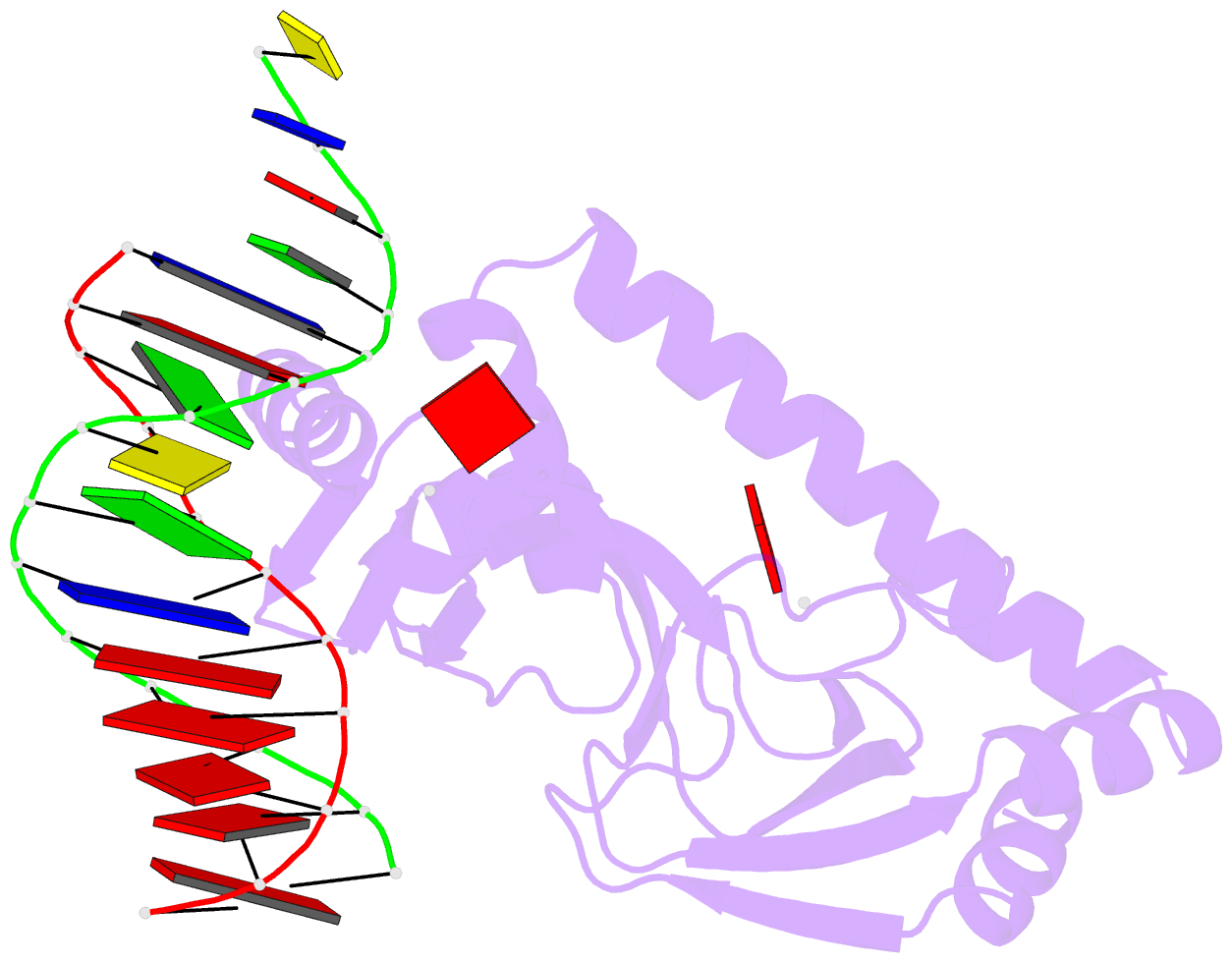
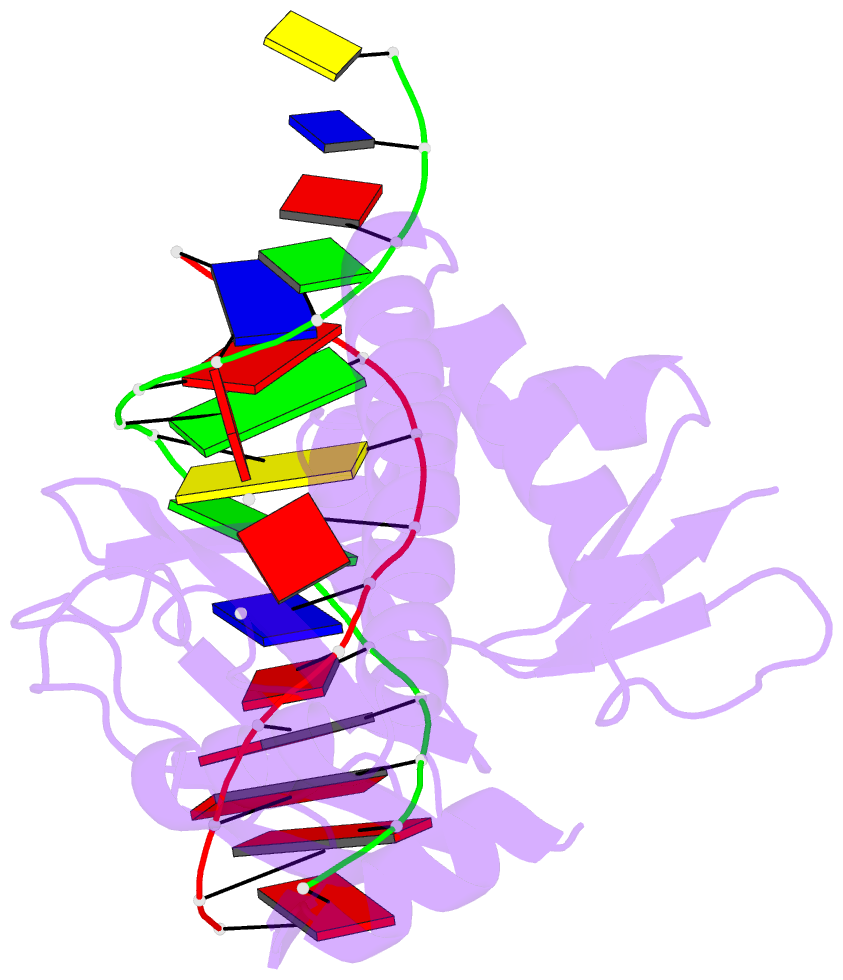
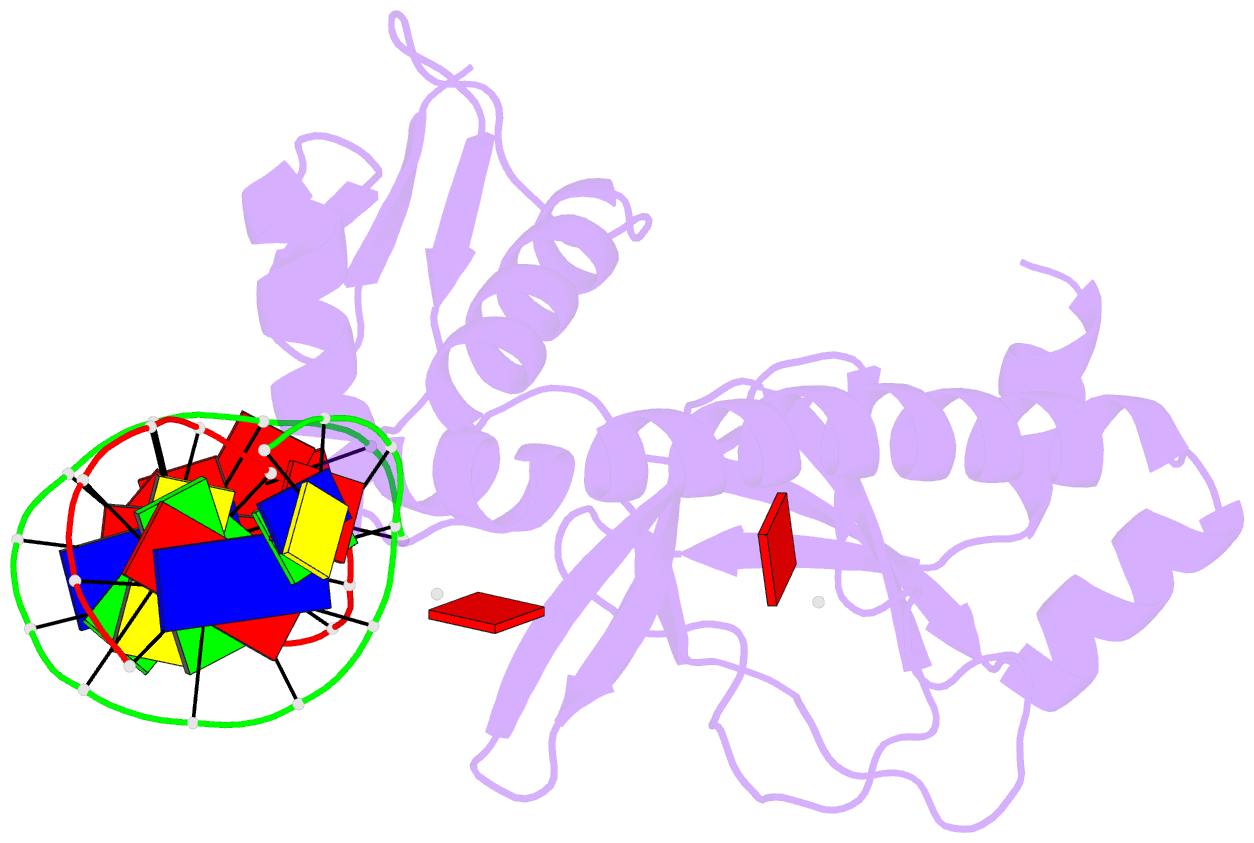
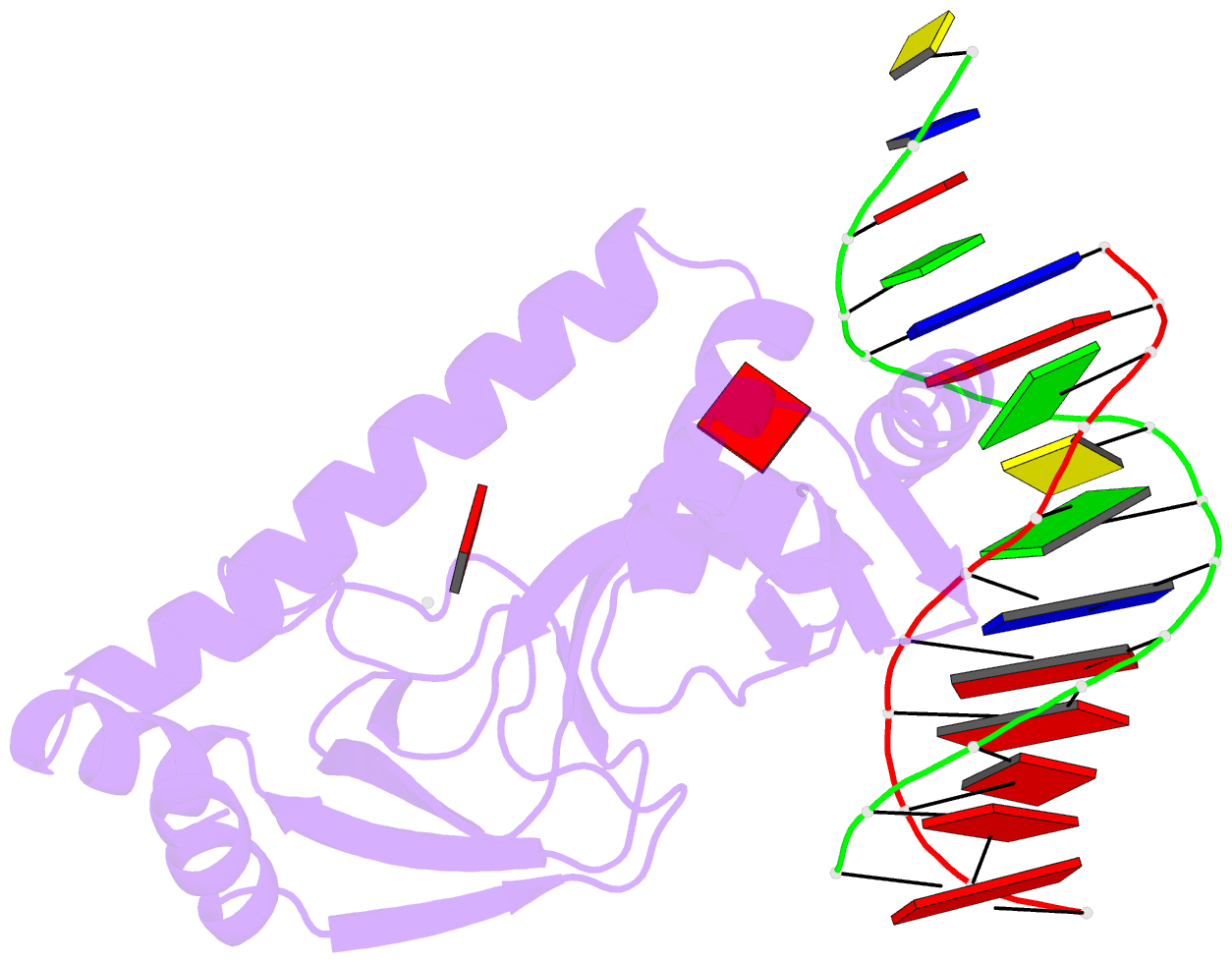
- Protein-DNA recognition and DNA deformation revealed in crystal structures of cap-DNA complexes
- Reference
- Chen S, Gunasekera A, Zhang X, Kunkel TA, Ebright RH, Berman HM (2001): "Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex: Alteration of DNA Binding Specificity Through Alteration of DNA Kinking." J.Mol.Biol., 314, 75-82. doi: 10.1006/jmbi.2001.5090.
- Abstract
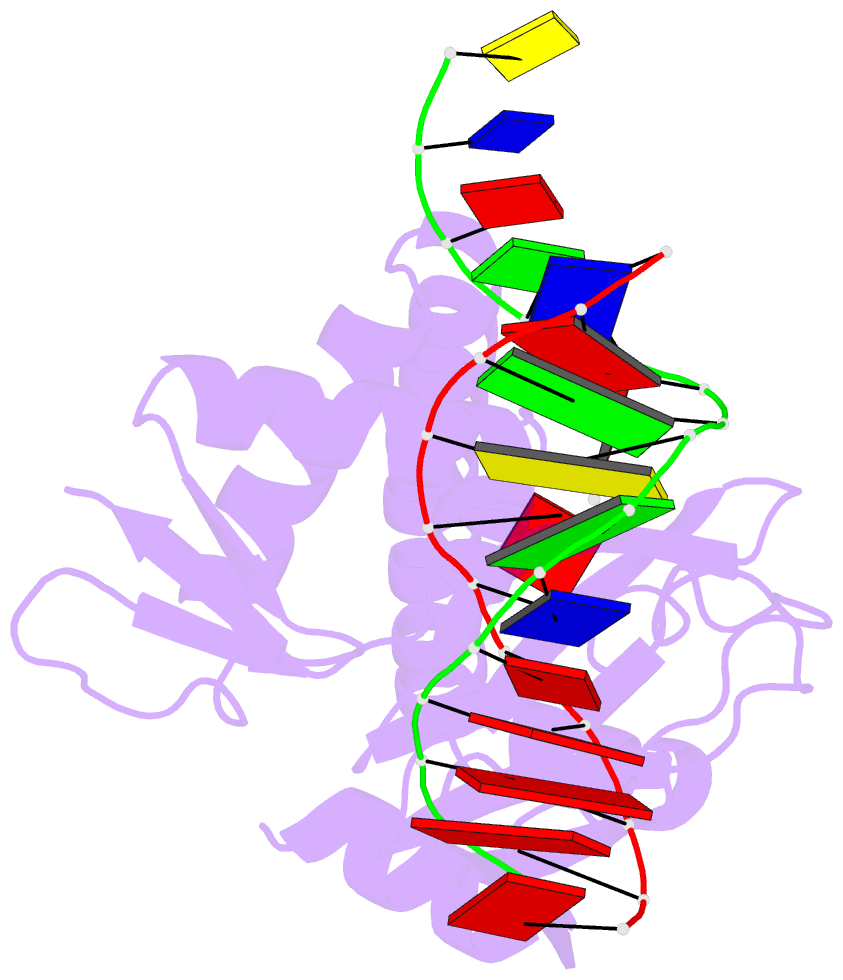
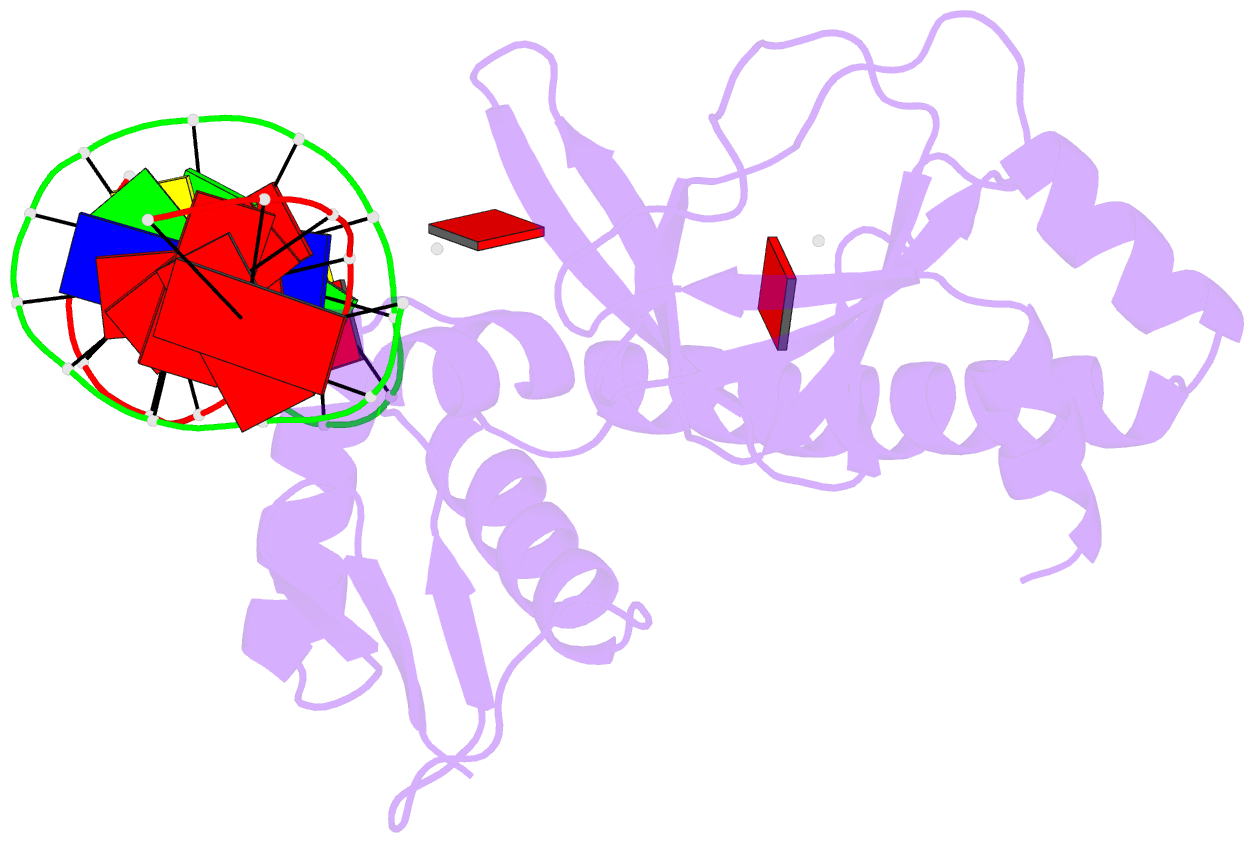
- The catabolite activator protein (CAP) sharply bends DNA in the CAP-DNA complex, introducing a DNA kink, with a roll angle of approximately 40 degrees and a twist angle of approximately 20 degrees, between positions 6 and 7 of the DNA half-site, 5'-A(1)A(2)A(3)T(4)G(5)T(6)G(7)A(8)T(9)C(10)T(11)-3' ("primary kink"). CAP recognizes the base-pair immediately 5' to the primary-kink site, T:A(6), through an "indirect-readout" mechanism involving sequence effects on the energetics of primary-kink formation. CAP recognizes the base-pair immediately 3' to the primary-kink site, G:C(7), through a "direct-readout" mechanism involving formation of a hydrogen bond between Glu181 of CAP and G:C(7). Here, we report that substitution of the carboxylate side-chain of Glu181 of CAP by the one-methylene-group-shorter carboxylate side-chain of Asp changes DNA binding specificity at position 6 of the DNA half site, changing specificity for T:A(6) to specificity for C:G(6), and we report a crystallographic analysis defining the structural basis of the change in specificity. The Glu181-->Asp substitution eliminates the primary kink and thus eliminates indirect-readout-based specificity for T:A(6). The Glu181-->Asp substitution does not eliminate hydrogen-bond formation with G:C(7), and thus does not eliminate direct-readout-based specificity for G:C(7).