Summary information and primary citation
- PDB-id
- 1p6v; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (3.2 Å)
- Summary
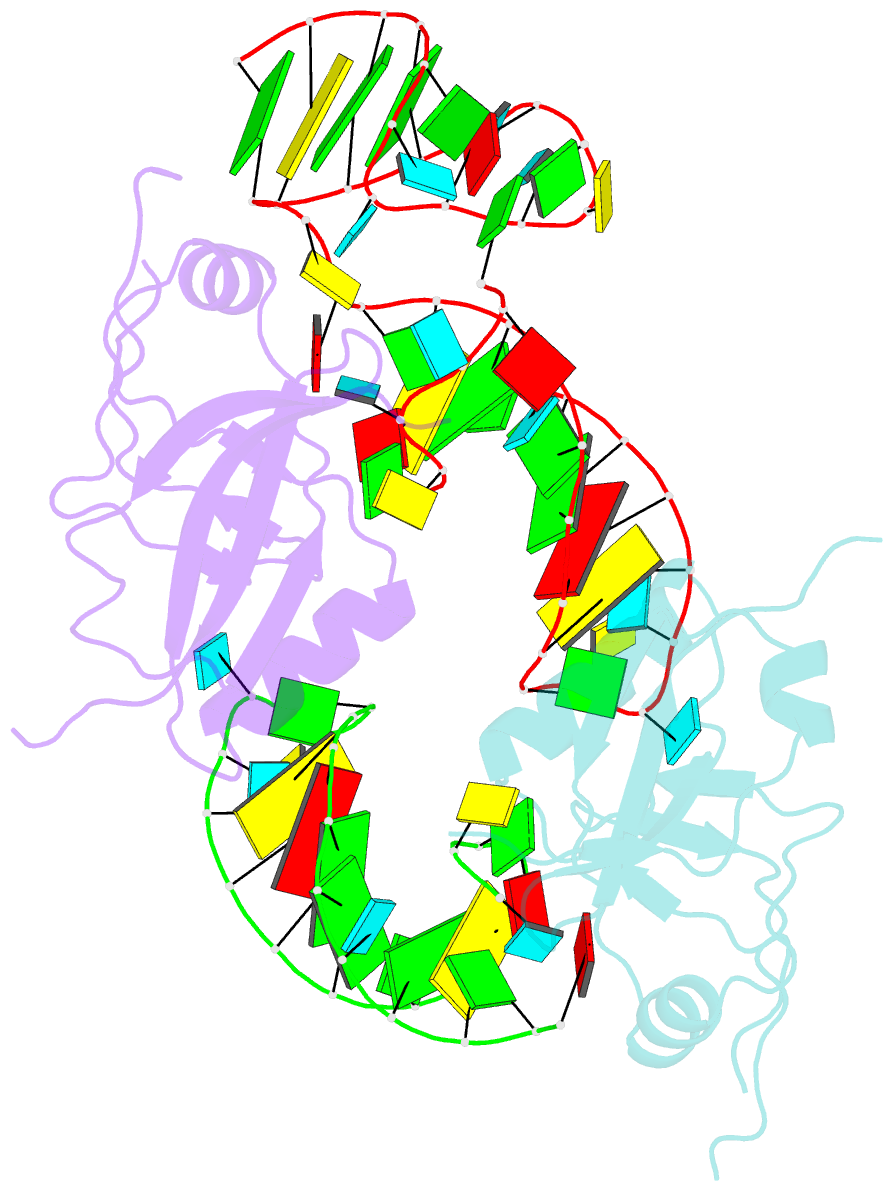
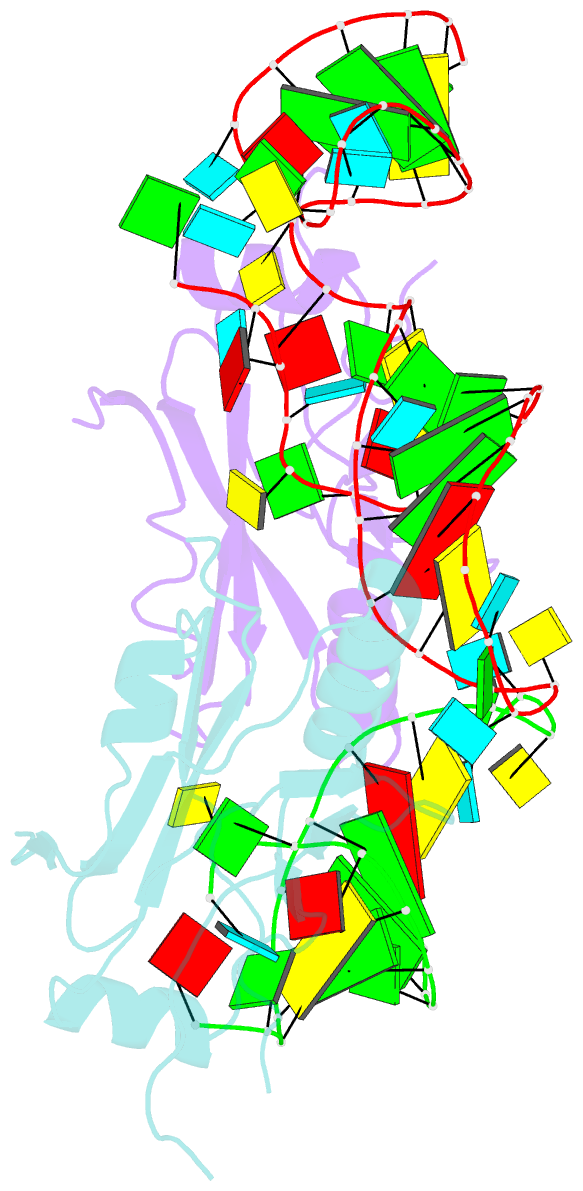
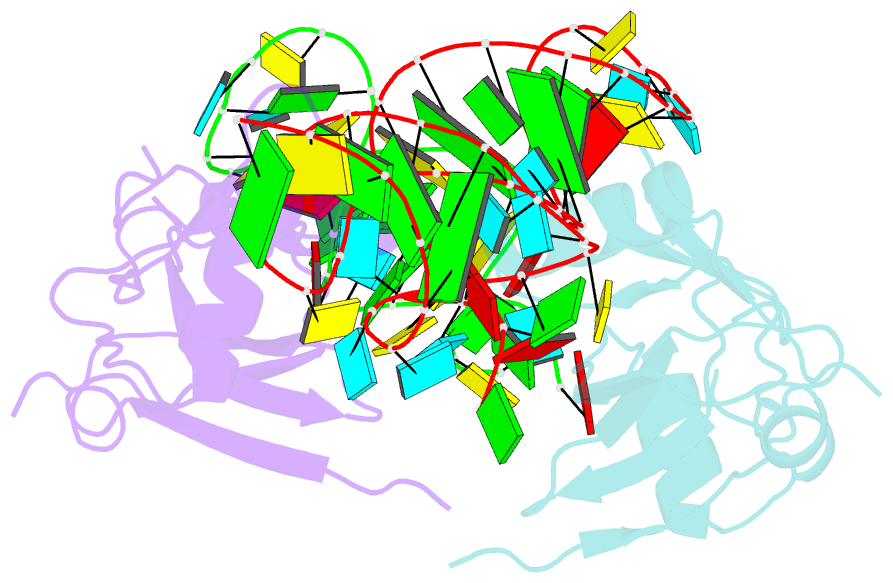
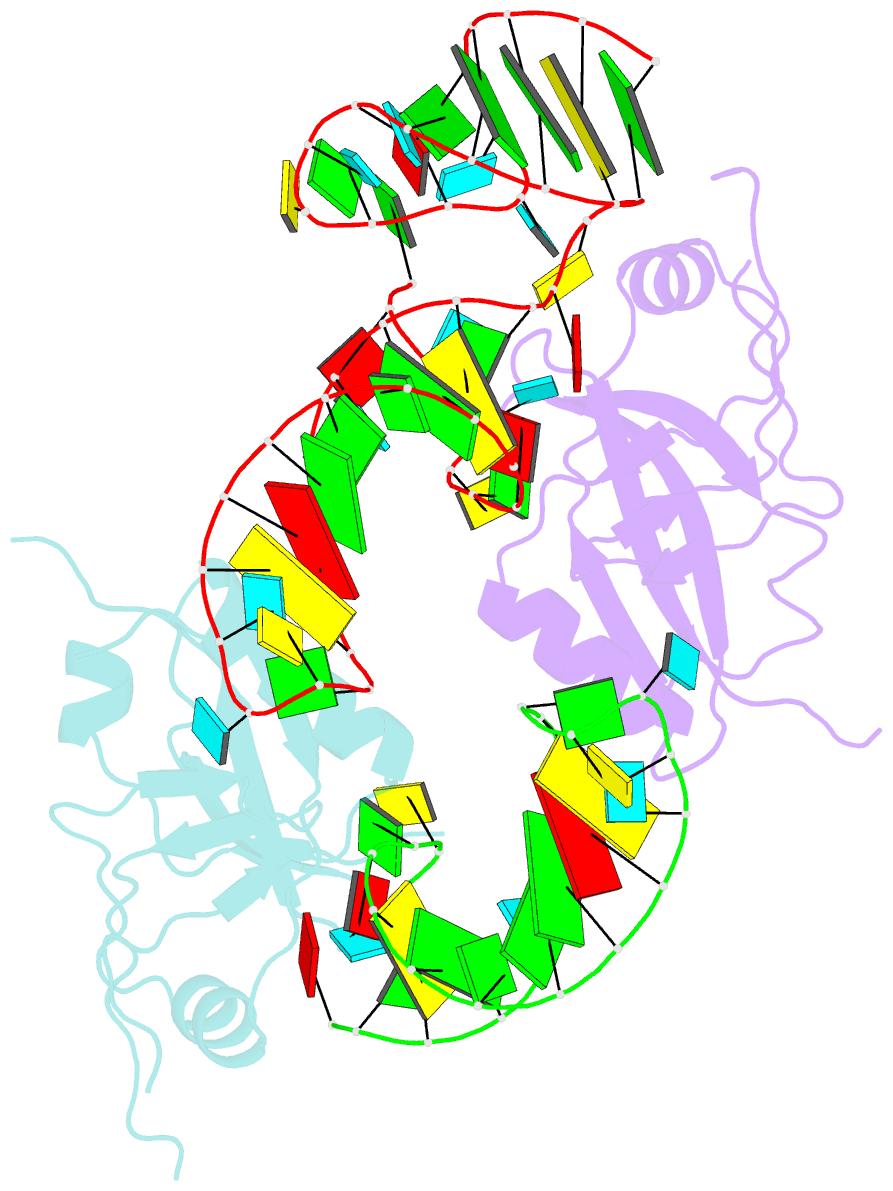
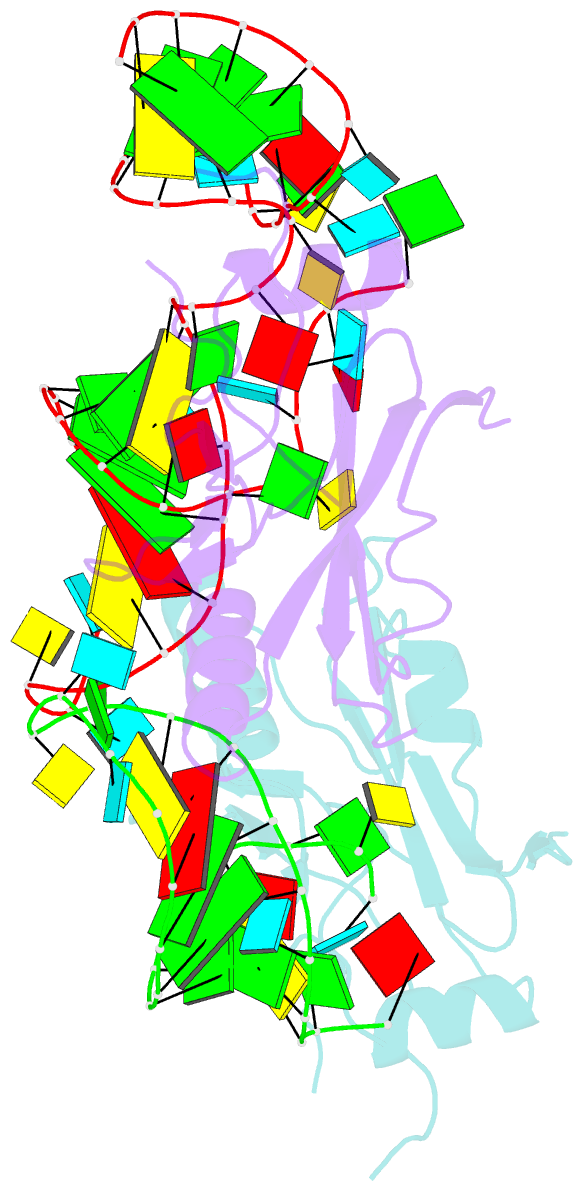
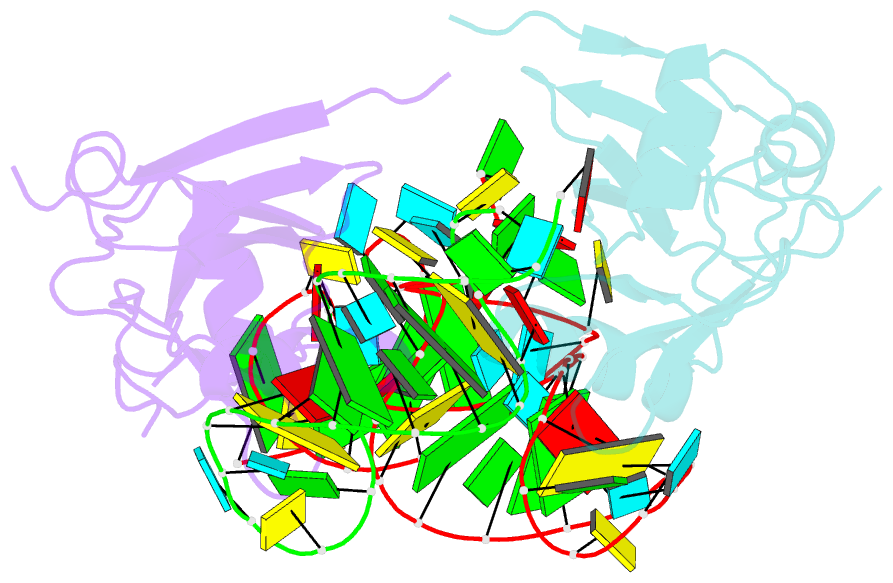
- Crystal structure of the trna domain of transfer-messenger RNA in complex with smpb
- Reference
- Gutmann S, Haebel PW, Metzinger L, Sutter M, Felden B, Ban N (2003): "Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB." Nature, 424, 699-703. doi: 10.1038/nature01831.
- Abstract
- Accurate translation of genetic information into protein sequence depends on complete messenger RNA molecules. Truncated mRNAs cause synthesis of defective proteins, and arrest ribosomes at the end of their incomplete message. In bacteria, a hybrid RNA molecule that combines the functions of both transfer and messenger RNAs (called tmRNA) rescues stalled ribosomes, and targets aberrant, partially synthesized, proteins for proteolytic degradation. Here we report the 3.2-A-resolution structure of the tRNA-like domain of tmRNA (tmRNA(Delta)) in complex with small protein B (SmpB), a protein essential for biological functions of tmRNA. We find that the flexible RNA molecule adopts an open L-shaped conformation and SmpB binds to its elbow region, stabilizing the single-stranded D-loop in an extended conformation. The most striking feature of the structure of tmRNA(Delta) is a 90 degrees rotation of the TPsiC-arm around the helical axis. Owing to this unusual conformation, the SmpB-tmRNA(Delta) complex positioned into the A-site of the ribosome orients SmpB towards the small ribosomal subunit, and directs tmRNA towards the elongation-factor binding region of the ribosome. On the basis of this structure, we propose a model for the binding of tmRNA on the ribosome.