Summary information and primary citation
- PDB-id
- 1pdn; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- gene regulation-DNA
- Method
- X-ray (2.5 Å)
- Summary
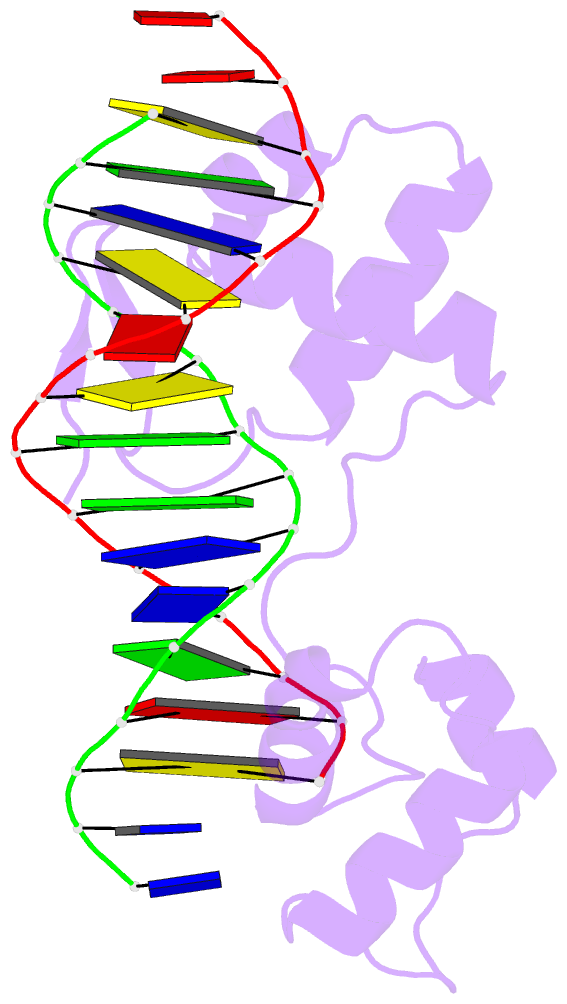
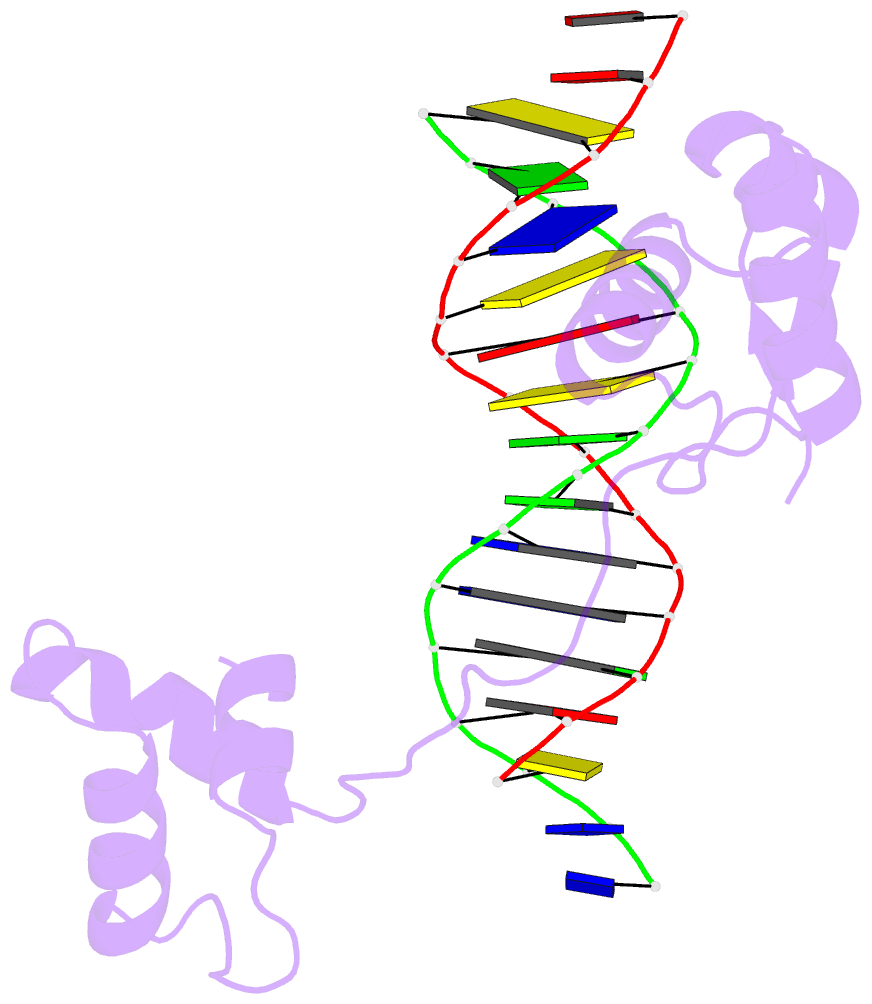
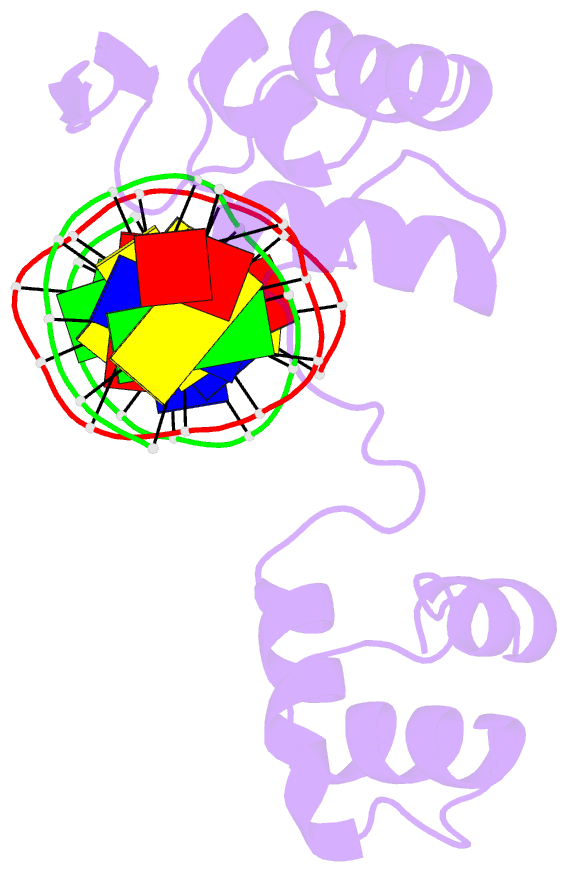
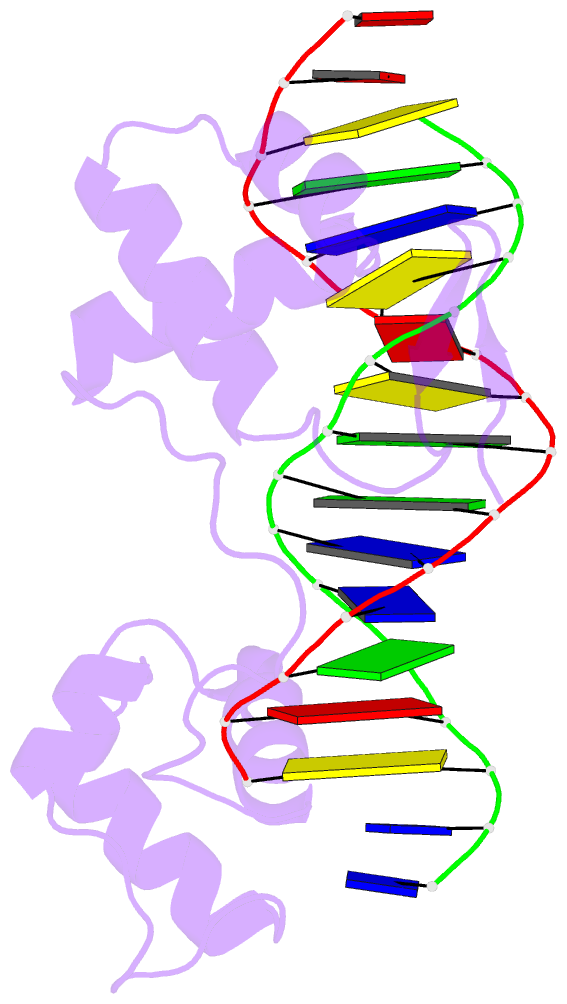
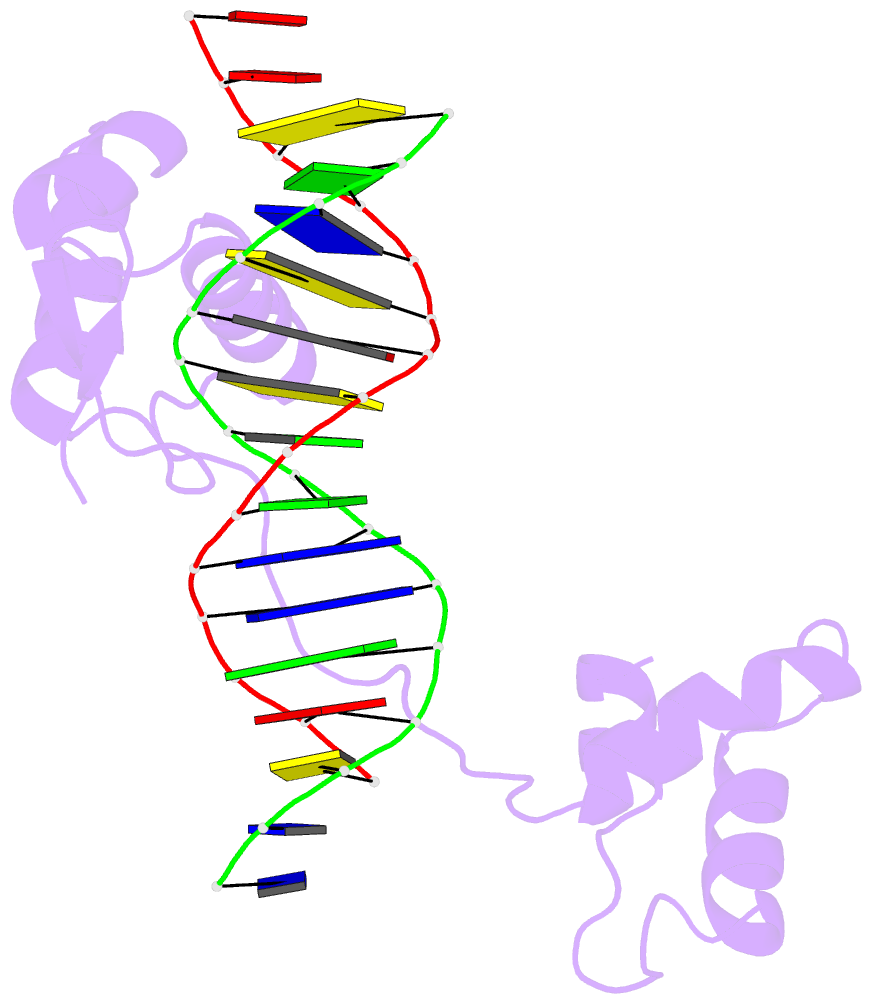
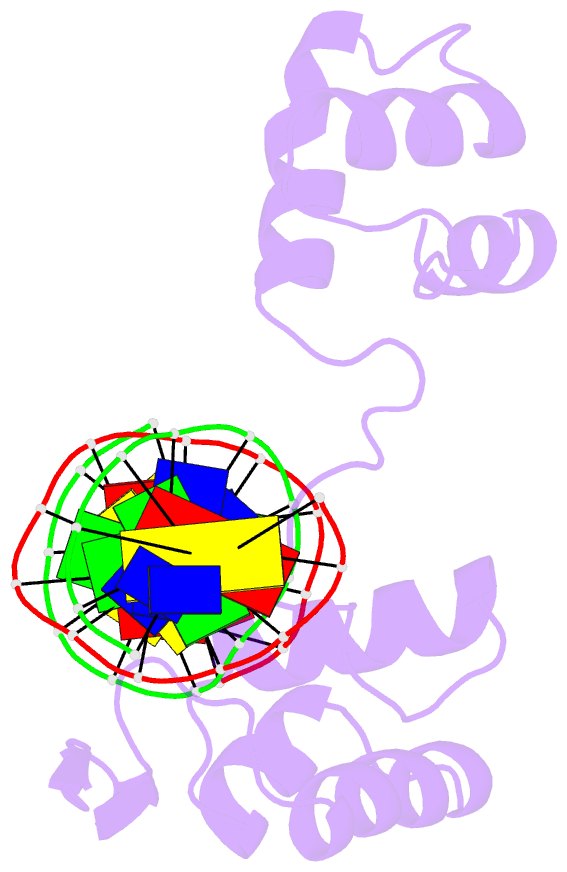
- Crystal structure of a paired domain-DNA complex at 2.5 angstroms resolution reveals structural basis for pax developmental mutations
- Reference
- Xu W, Rould MA, Jun S, Desplan C, Pabo CO (1995): "Crystal structure of a paired domain-DNA complex at 2.5 A resolution reveals structural basis for Pax developmental mutations." Cell(Cambridge,Mass.), 80, 639-650. doi: 10.1016/0092-8674(95)90518-9.
- Abstract
- The 2.5 A resolution structure of a cocrystal containing the paired domain from the Drosophila paired (prd) protein and a 15 bp site shows structurally independent N-terminal and C-terminal subdomains. Each of these domains contains a helical region resembling the homeodomain and the Hin recombinase. The N-terminal domain makes extensive DNA contacts, using a novel beta turn motif that binds in the minor groove and a helix-turn-helix unit with a docking arrangement surprisingly similar to that of the lambda repressor. The C-terminal domain is not essential for prd binding and does not contact the optimized site. All known developmental missense mutations in the paired box of mammalian Pax genes map to the N-terminal subdomain, and most of them are found at the protein-DNA interface.