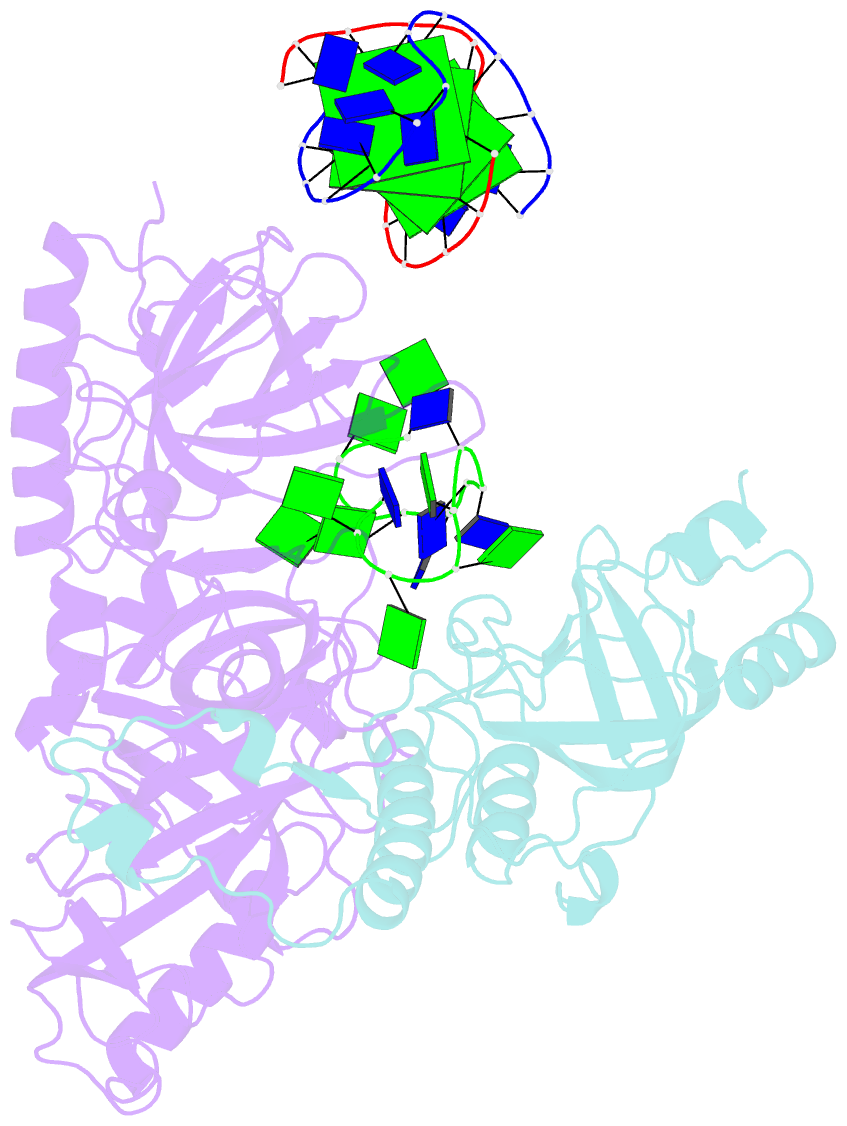
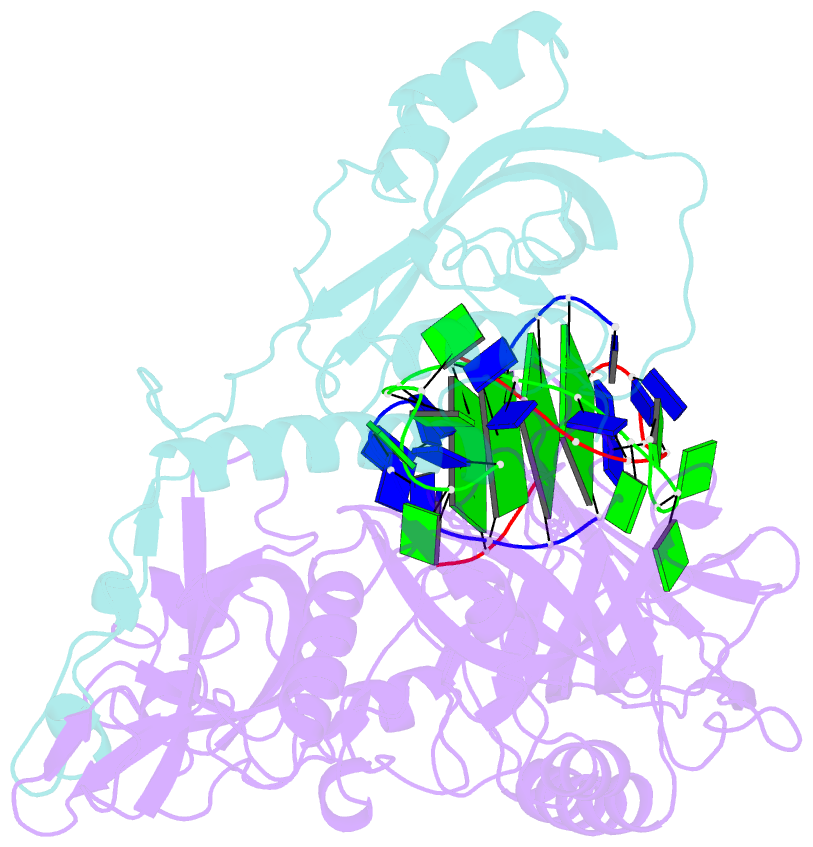
Summary information and primary citation
- PDB-id
- 1ph1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.51 Å)
- Summary
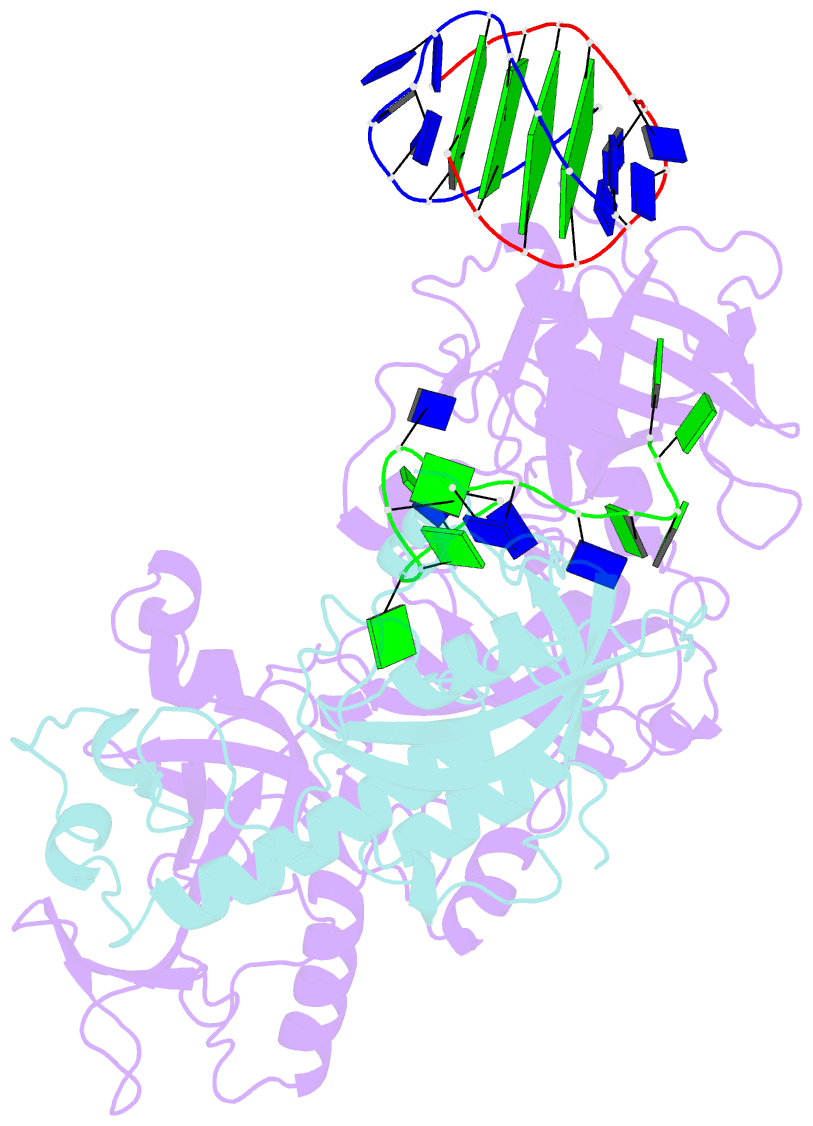
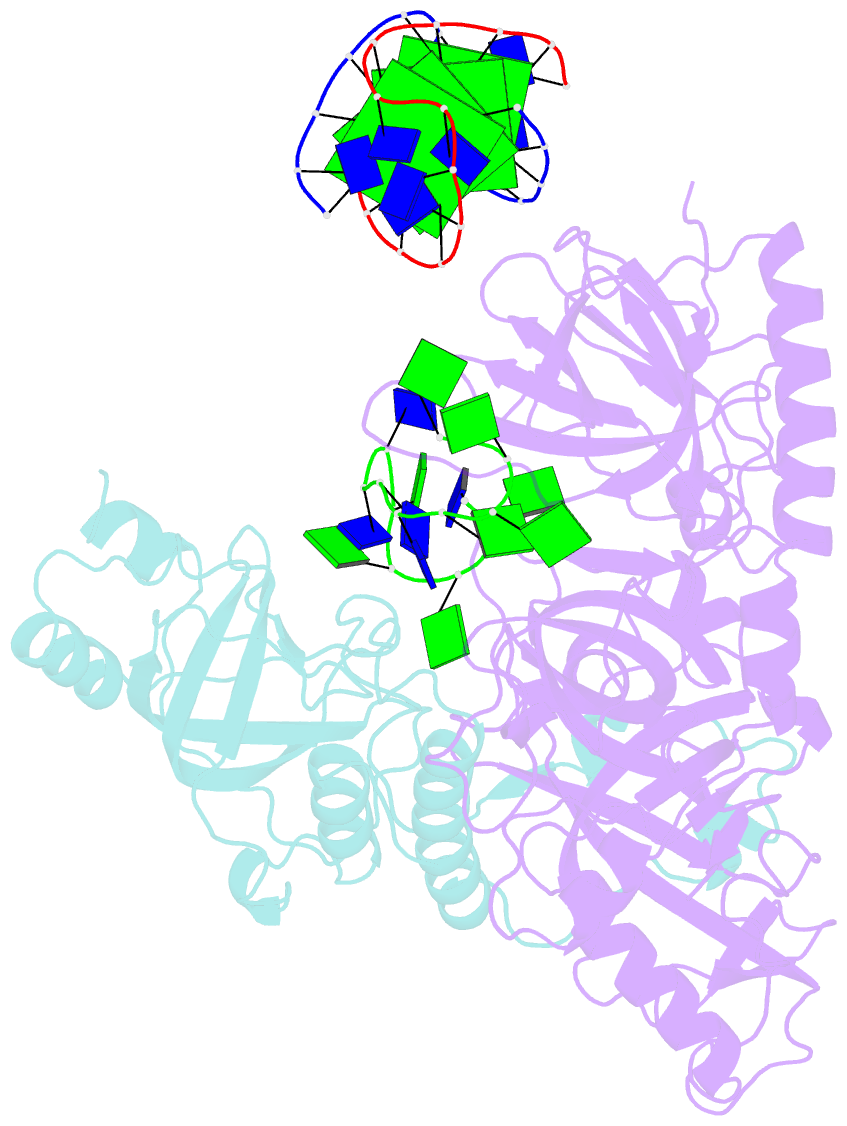
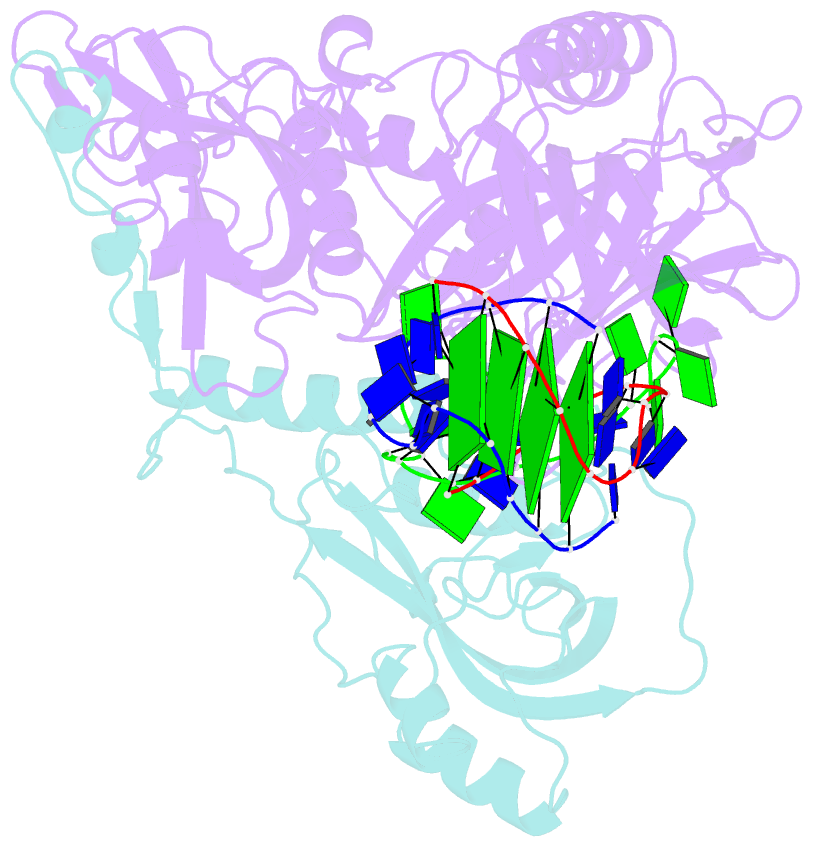
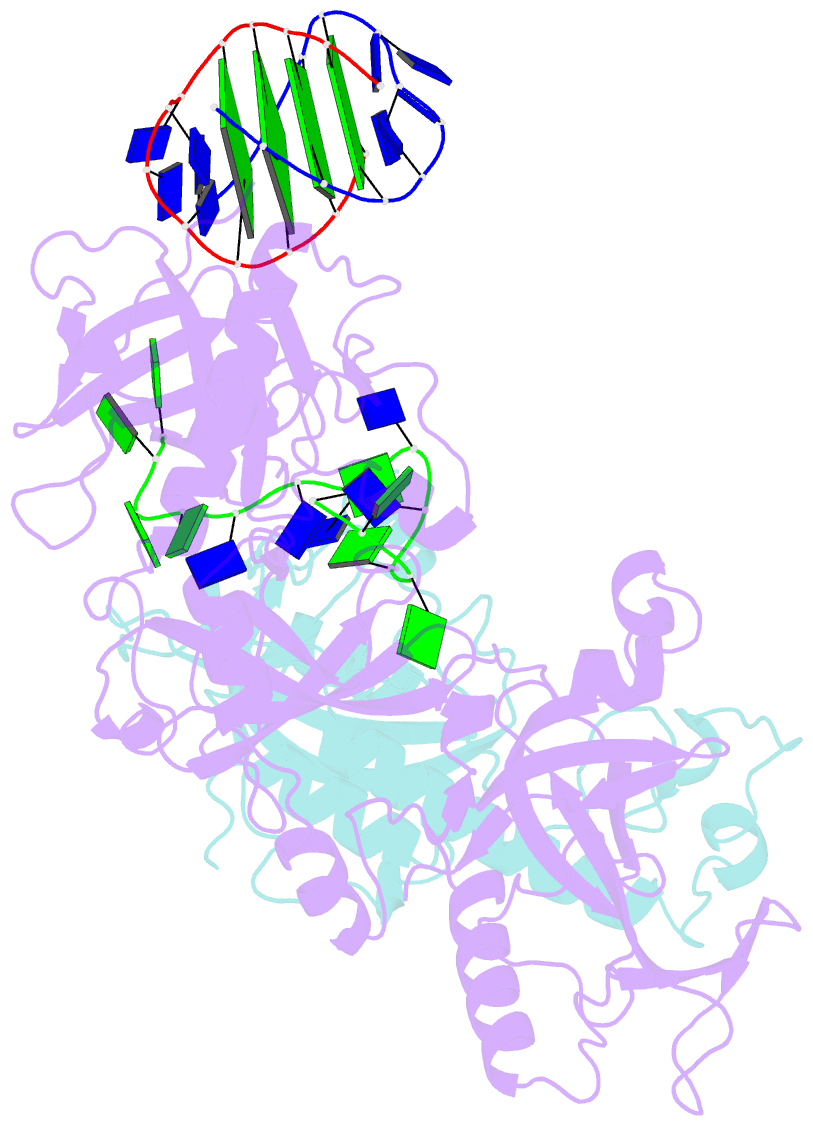
- Crystal structure of the oxytricha nova telomere end-binding protein complexed with noncognate ssDNA ggggttttggggt
- Reference
- Theobald DL, Schultz SC (2003): "Nucleotide Shuffling and Ssdna Recognition in Oxytricha Nova Telomere End-Binding Protein Complexes." Embo J., 22, 4314-4324. doi: 10.1093/emboj/cdg415.
- Abstract
- Sequence-specific protein recognition of single-stranded nucleic acids is critical for many fundamental cellular processes, such as DNA replication, DNA repair, transcription, translation, recombination, apoptosis and telomere maintenance. To explore the mechanisms of sequence-specific ssDNA recognition, we determined the crystal structures of 10 different non-cognate ssDNAs complexed with the Oxytricha nova telomere end-binding protein (OnTEBP) and evaluated their corresponding binding affinities (PDB ID codes 1PH1-1PH9 and 1PHJ). The thermodynamic and structural effects of these sequence perturbations could not have been predicted based solely upon the cognate structure. OnTEBP accommodates non-cognate nucleotides by both subtle adjustments and surprisingly large structural rearrangements in the ssDNA. In two complexes containing ssDNA intermediates that occur during telomere extension by telomerase, entire nucleotides are expelled from the complex. Concurrently, the sequence register of the ssDNA shifts to re-establish a more cognate-like pattern. This phenomenon, termed nucleotide shuffling, may be of general importance in protein recognition of single-stranded nucleic acids. This set of structural and thermodynamic data highlights a fundamental difference between protein recognition of ssDNA versus dsDNA.