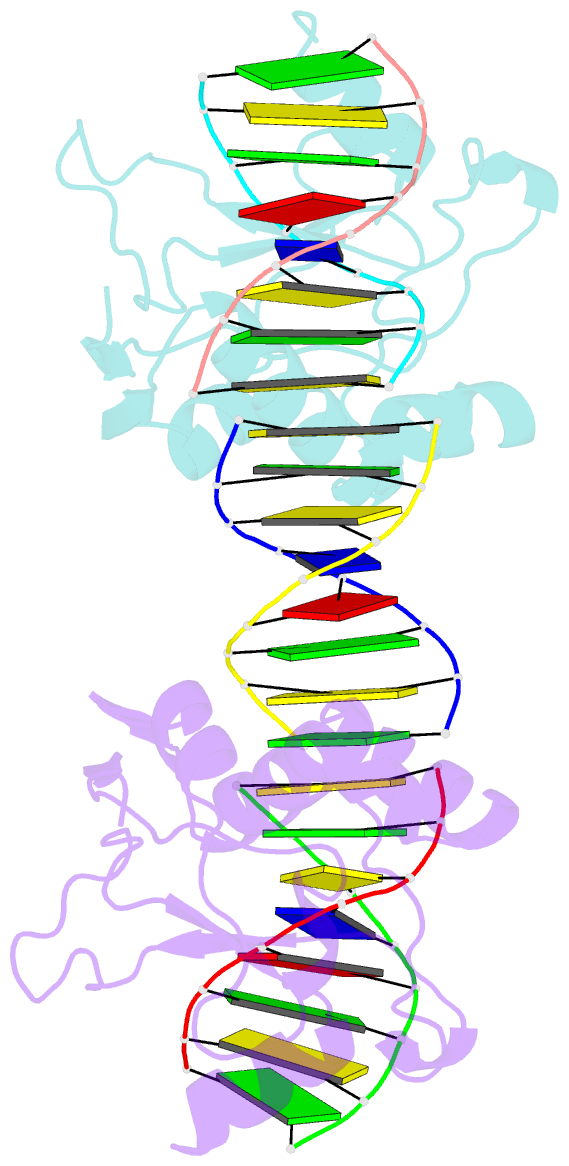
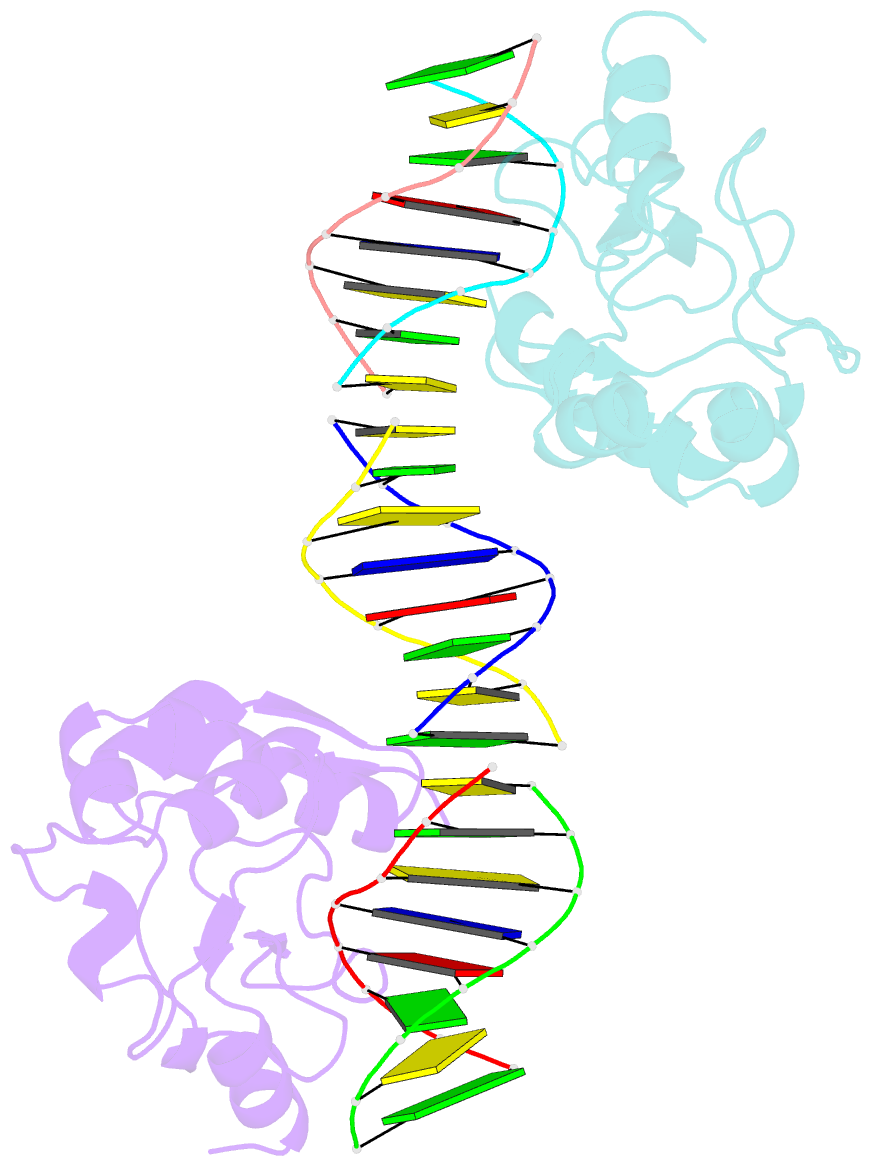
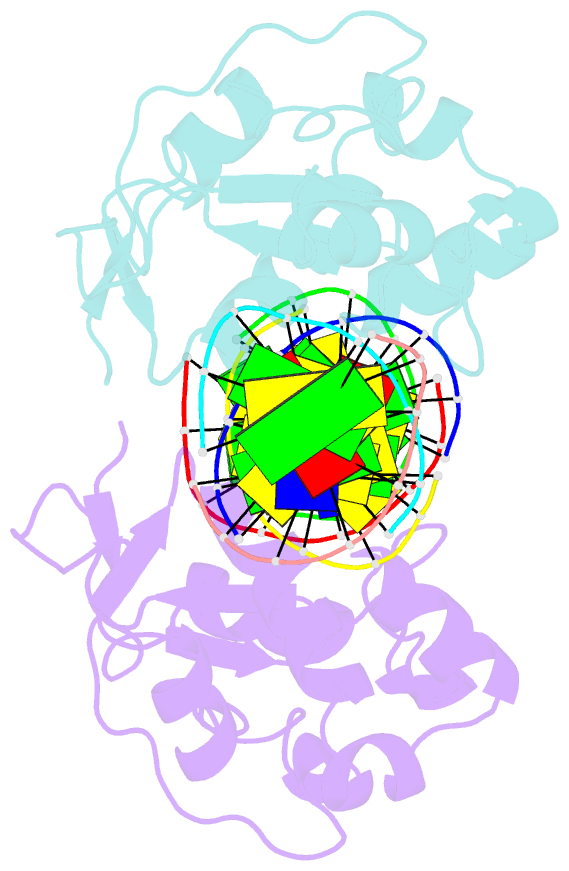
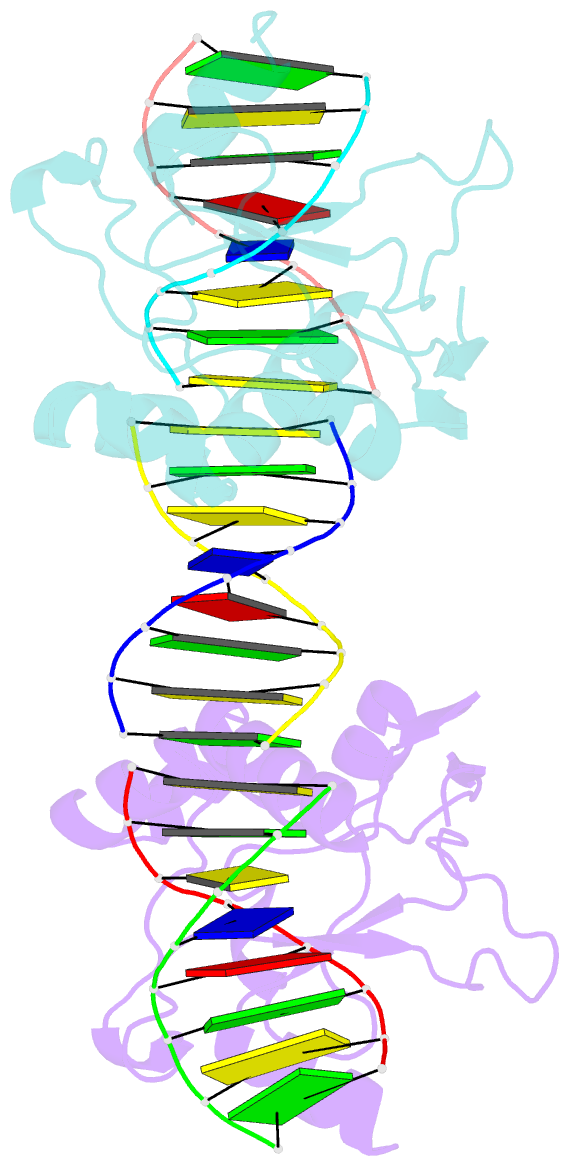
Summary information and primary citation
- PDB-id
- 1pt3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.5 Å)
- Summary
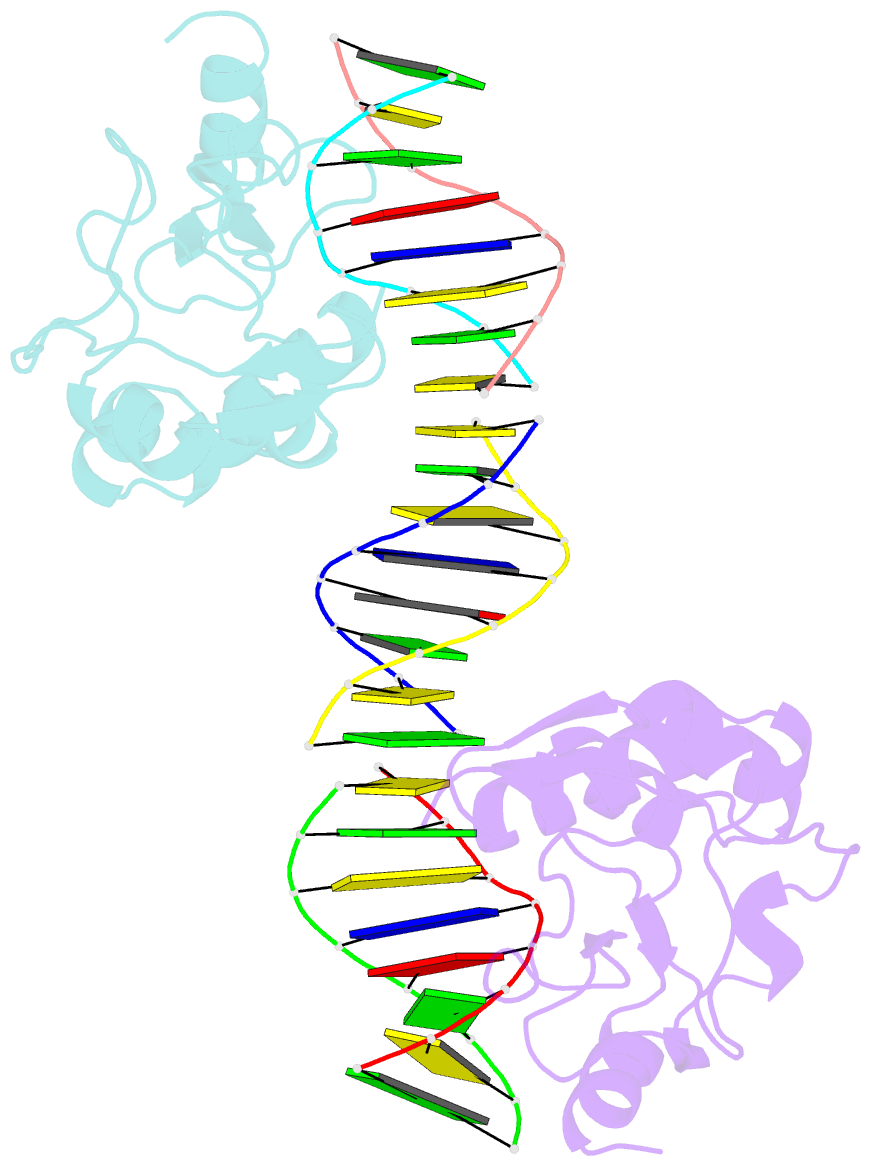
- Crystal structures of nuclease-cole7 complexed with octamer DNA
- Reference
- Hsia KC, Chak KF, Liang PH, Cheng YS, Ku WY, Yuan HS (2004): "DNA binding and degradation by the HNH protein ColE7." STRUCTURE, 12, 205-214. doi: 10.1016/S0969-2126(04)00006-1.
- Abstract
- The bacterial toxin ColE7 bears an HNH motif which has been identified in hundreds of prokaryotic and eukaryotic endonucleases, involved in DNA homing, restriction, repair, or chromosome degradation. The crystal structure of the nuclease domain of ColE7 in complex with a duplex DNA has been determined at 2.5 A resolution. The HNH motif is bound at the minor groove primarily to DNA phosphate groups at and beyond the 3' side of the scissile phosphate, with little interaction with ribose groups and bases. This result provides a structural basis for sugar- and sequence-independent DNA recognition and the inhibition mechanism by inhibitor Im7, which blocks the substrate binding site but not the active site. Structural comparison shows that two families of endonucleases bind and bend DNA in a similar way to that of the HNH ColE7, indicating that endonucleases containing a "betabetaalpha-metal" fold of active site possess a universal mode for protein-DNA interactions.