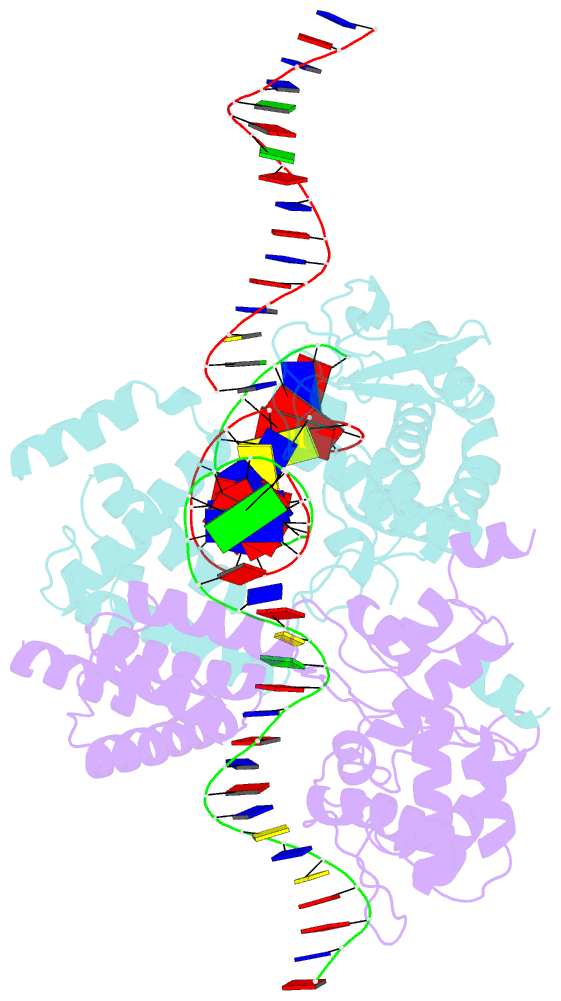
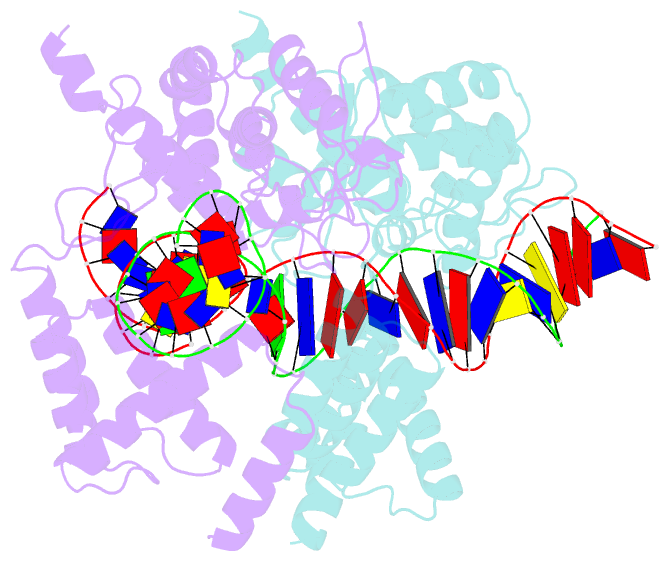
Summary information and primary citation
- PDB-id
- 1pvp; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- recombination-DNA
- Method
- X-ray (2.35 Å)
- Summary
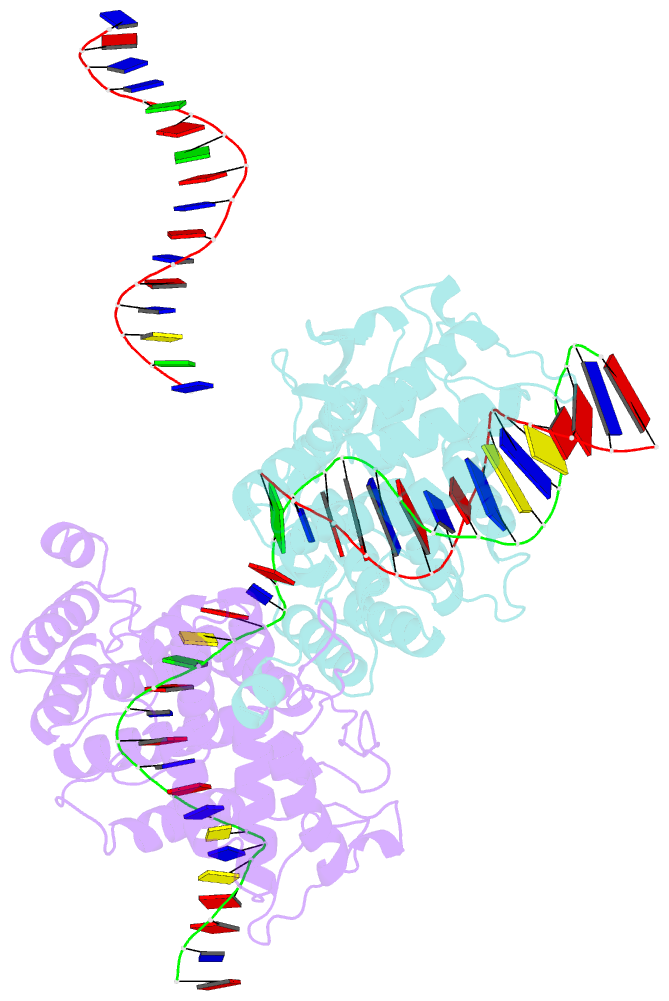
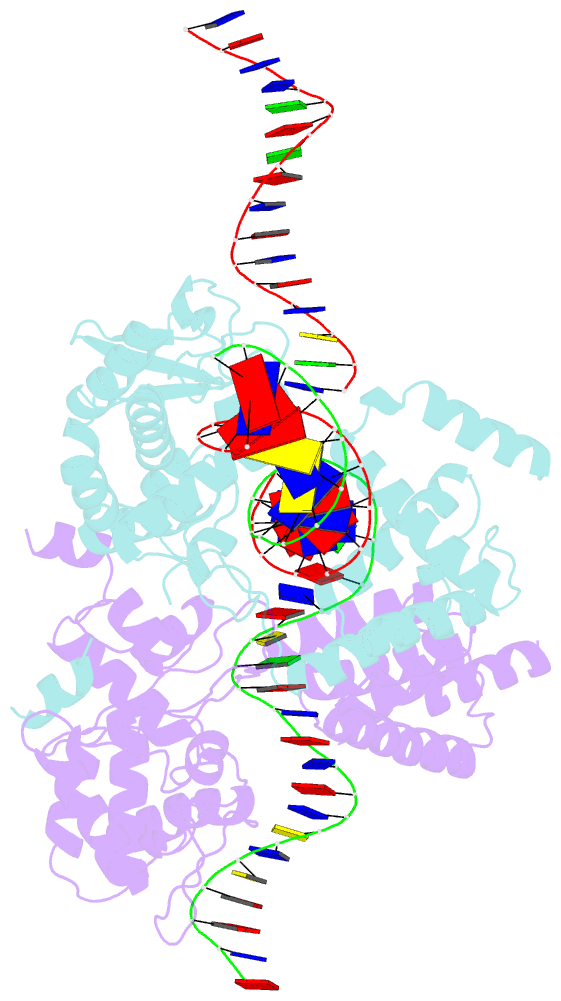
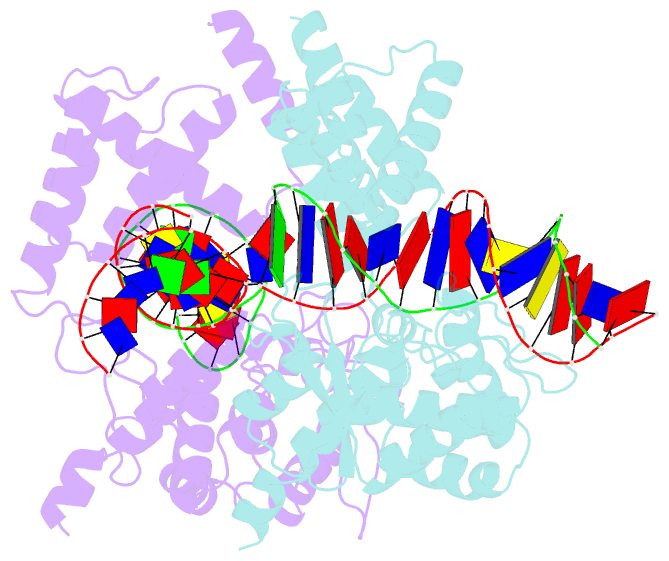
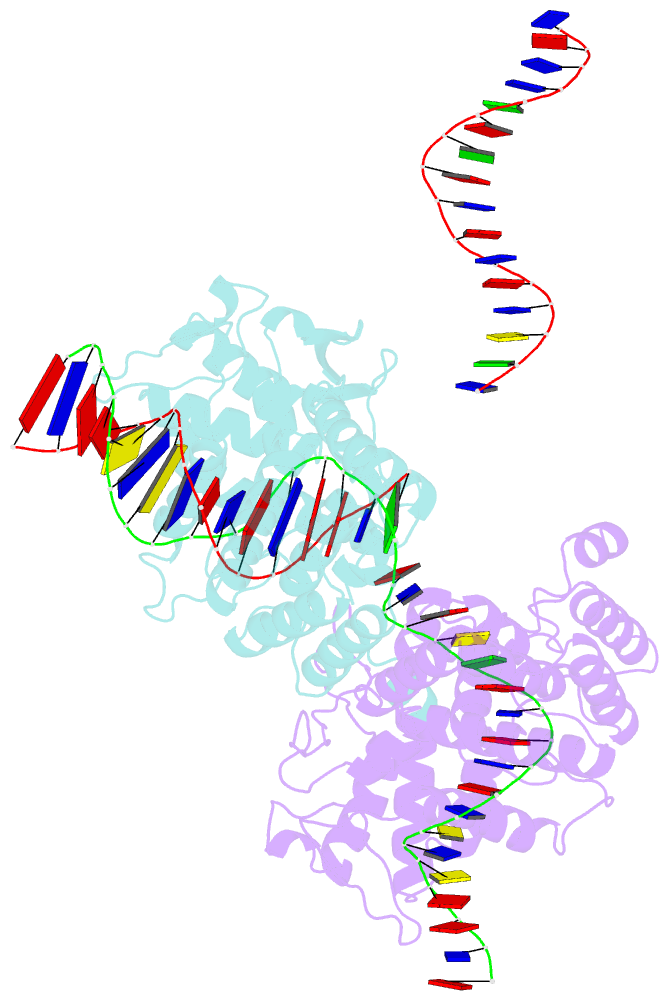
- Basis for a switch in substrate specificity: crystal structure of selected variant of cre site-specific recombinase, alshg bound to the engineered recognition site loxm7
- Reference
- Baldwin EP, Martin SS, Abel J, Gelato KA, Kim H, Schultz PG, Santoro SW (2003): "A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water." Chem.Biol., 10, 1085-1094. doi: 10.1016/j.chembiol.2003.10.015.
- Abstract
- The basis for the altered DNA specificities of two Cre recombinase variants, obtained by mutation and selection, was revealed by their cocrystal structures. The proteins share similar substitutions but differ in their preferences for the natural LoxP substrate and an engineered substrate that is inactive with wild-type Cre, LoxM7. One variant preferentially recombines LoxM7 and contacts the substituted bases through a hydrated network of novel interlocking protein-DNA contacts. The other variant recognizes both LoxP and LoxM7 utilizing the same DNA backbone contact but different base contacts, facilitated by an unexpected DNA shift. Assisted by water, novel interaction networks can arise from few protein substitutions, suggesting how new DNA binding specificities might evolve. The contributions of macromolecular plasticity and water networks in specific DNA recognition observed here present a challenge for predictive schemes.