Summary information and primary citation
- PDB-id
- 1q0t; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (3.1 Å)
- Summary
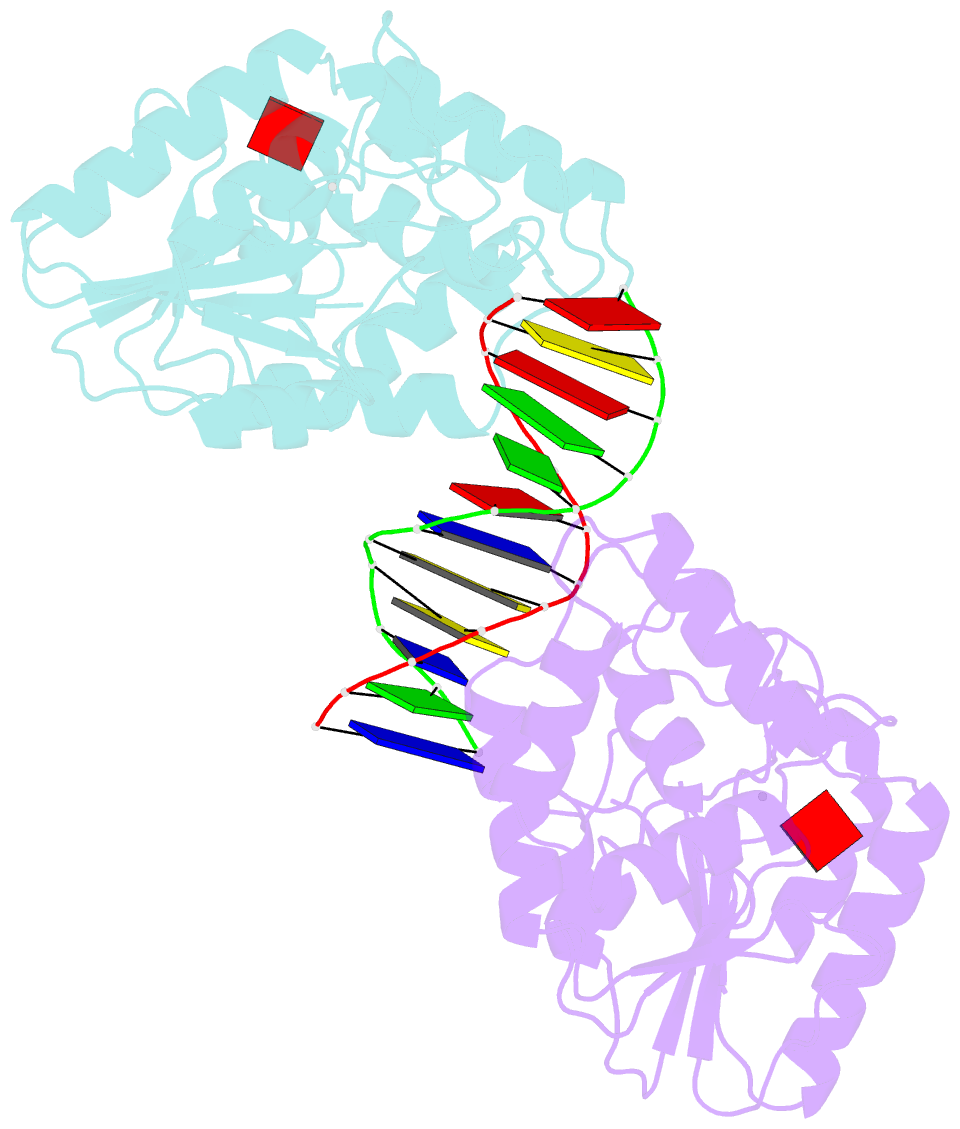
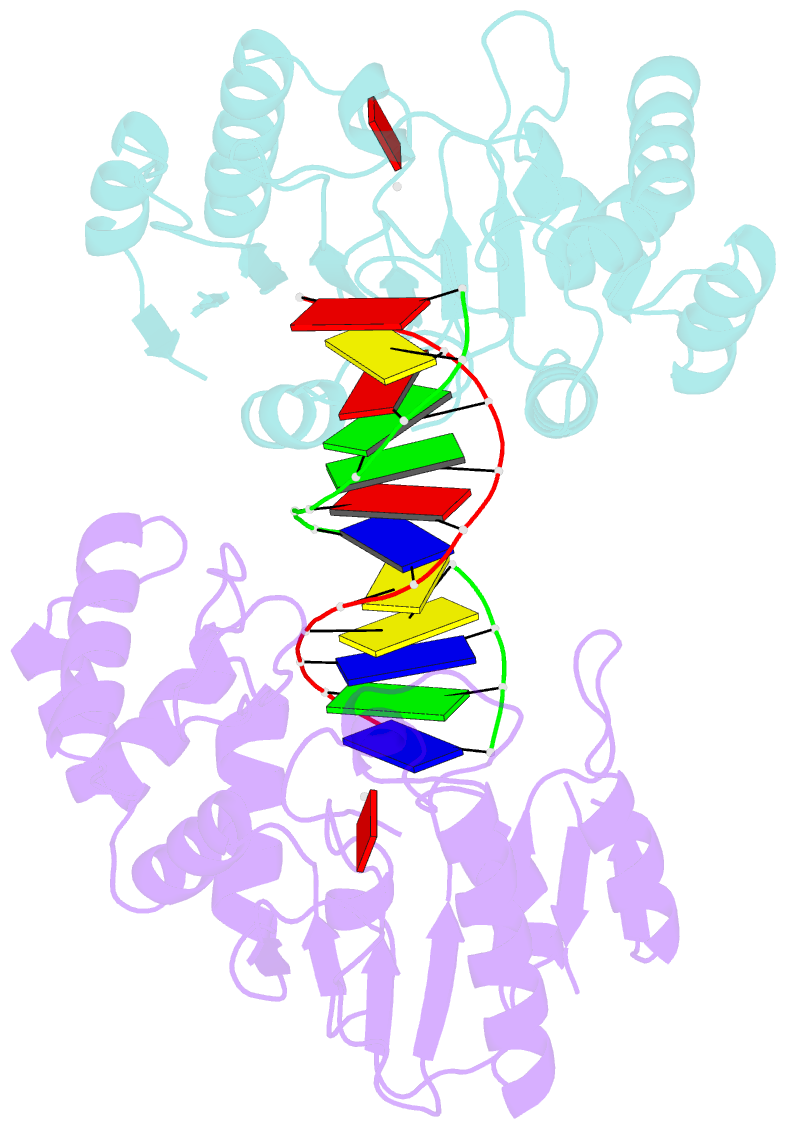
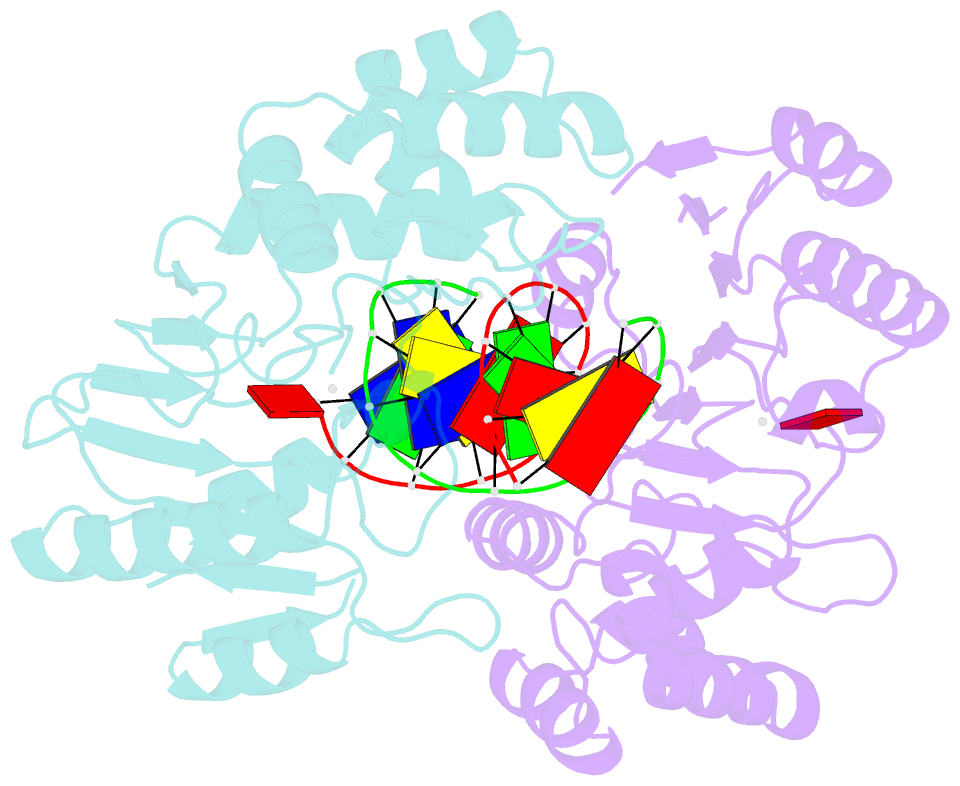
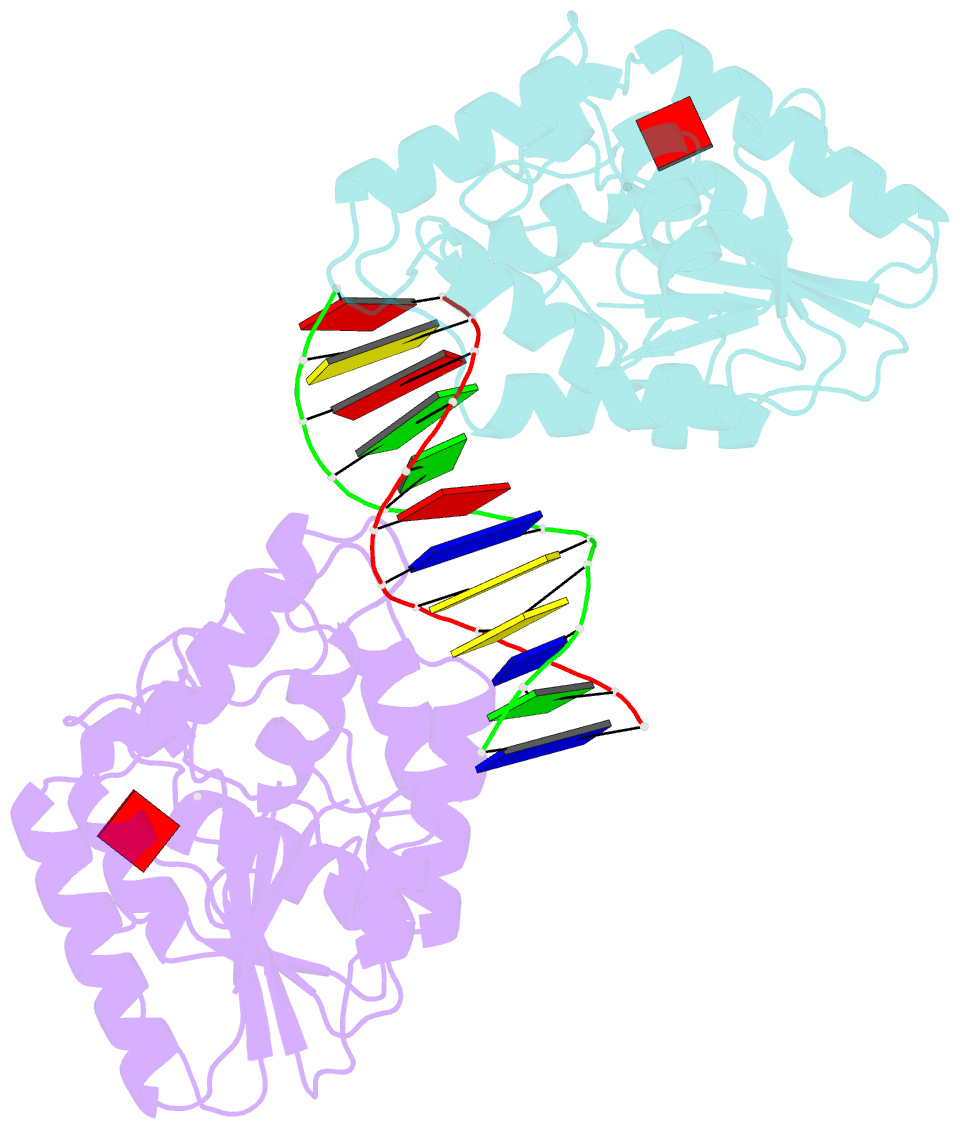
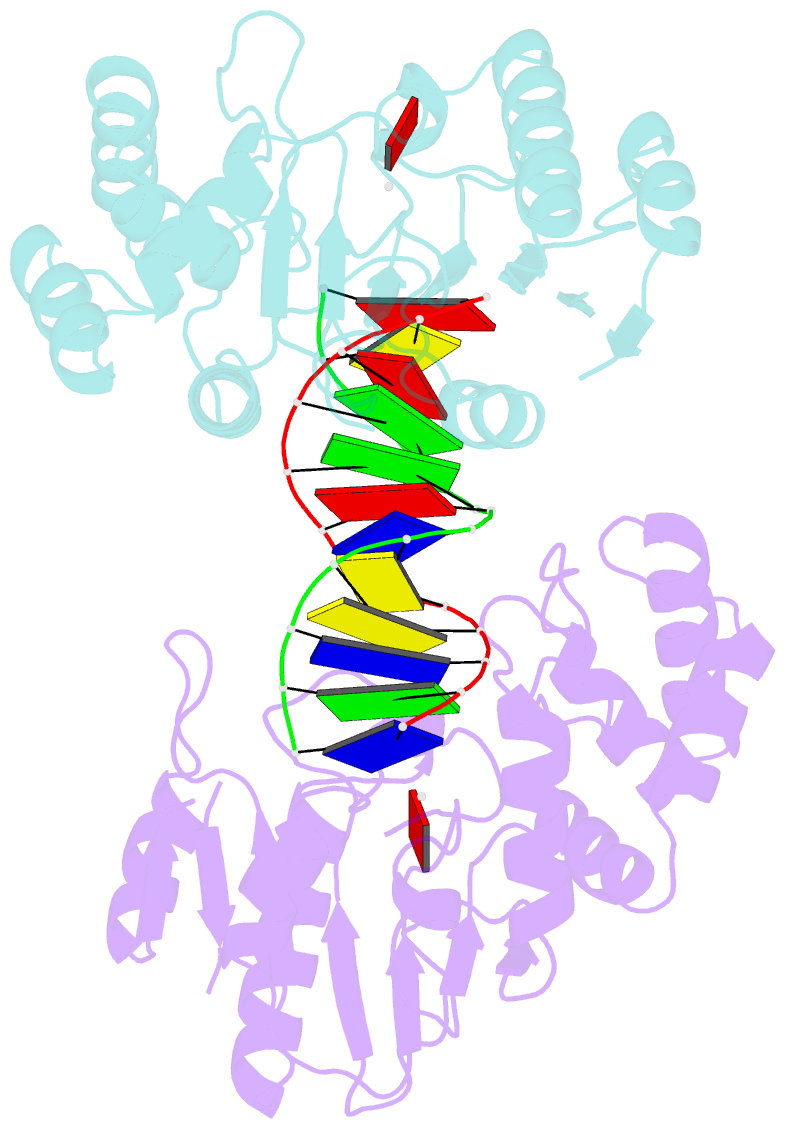
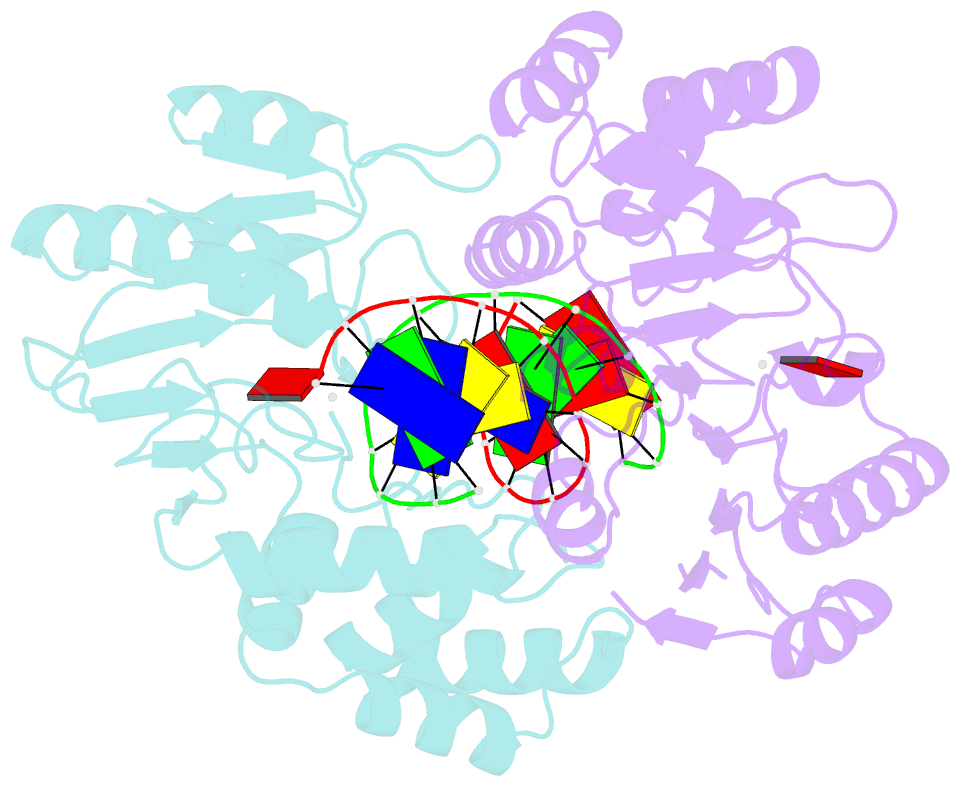
- Ternary structure of t4dam with adohcy and DNA
- Reference
- Yang Z, Horton JR, Zhou L, Zhang XJ, Dong A, Zhang X, Schlagman SL, Kossykh V, Hattman S, Cheng X (2003): "Structure of the bacteriophage T4 DNA adenine methyltransferase." Nat.Struct.Biol., 10, 849-855. doi: 10.1038/nsb973.
- Abstract
- DNA-adenine methylation at certain GATC sites plays a pivotal role in bacterial and phage gene expression as well as bacterial virulence. We report here the crystal structures of the bacteriophage T4Dam DNA adenine methyltransferase (MTase) in a binary complex with the methyl-donor product S-adenosyl-L-homocysteine (AdoHcy) and in a ternary complex with a synthetic 12-bp DNA duplex and AdoHcy. T4Dam contains two domains: a seven-stranded catalytic domain that harbors the binding site for AdoHcy and a DNA binding domain consisting of a five-helix bundle and a beta-hairpin that is conserved in the family of GATC-related MTase orthologs. Unexpectedly, the sequence-specific T4Dam bound to DNA in a nonspecific mode that contained two Dam monomers per synthetic duplex, even though the DNA contains a single GATC site. The ternary structure provides a rare snapshot of an enzyme poised for linear diffusion along the DNA.