Summary information and primary citation
- PDB-id
- 1qf6; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ligase-RNA
- Method
- X-ray (2.9 Å)
- Summary
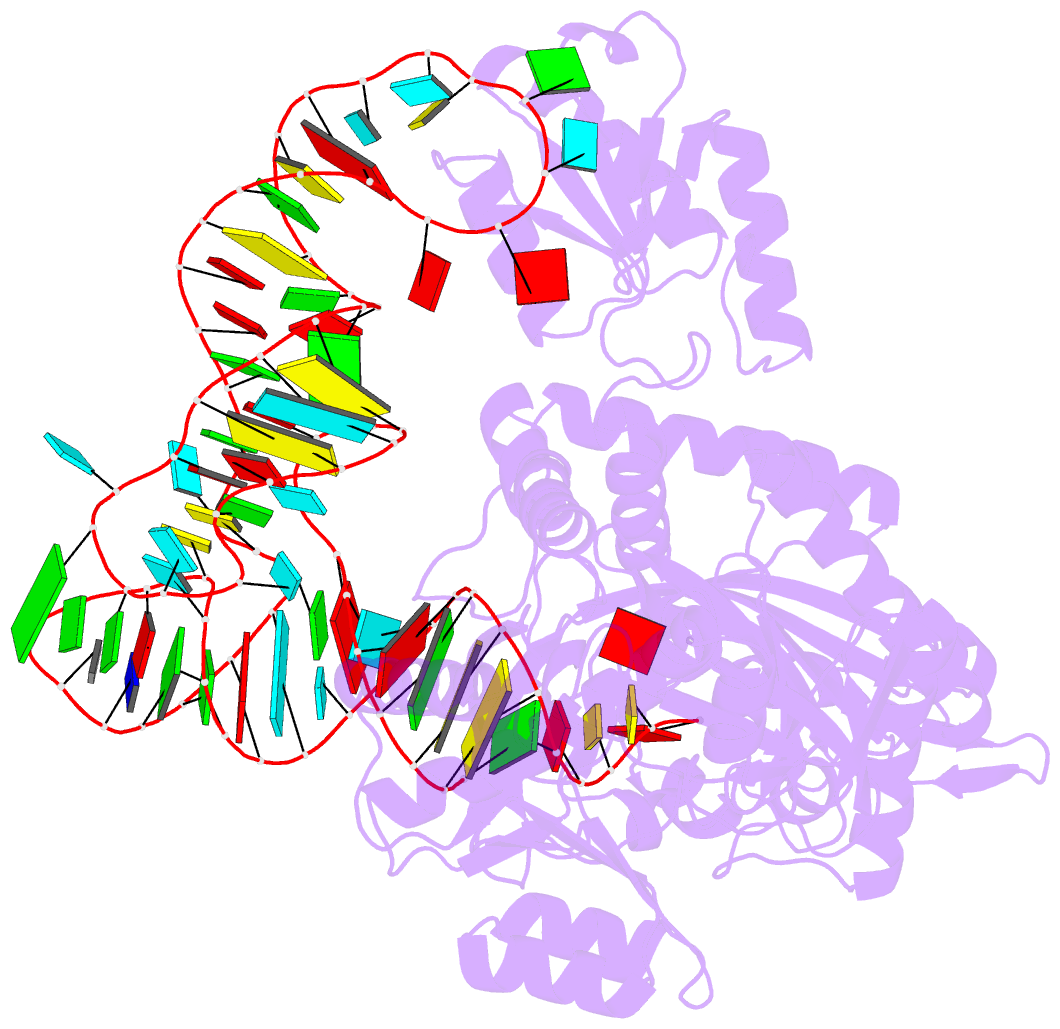
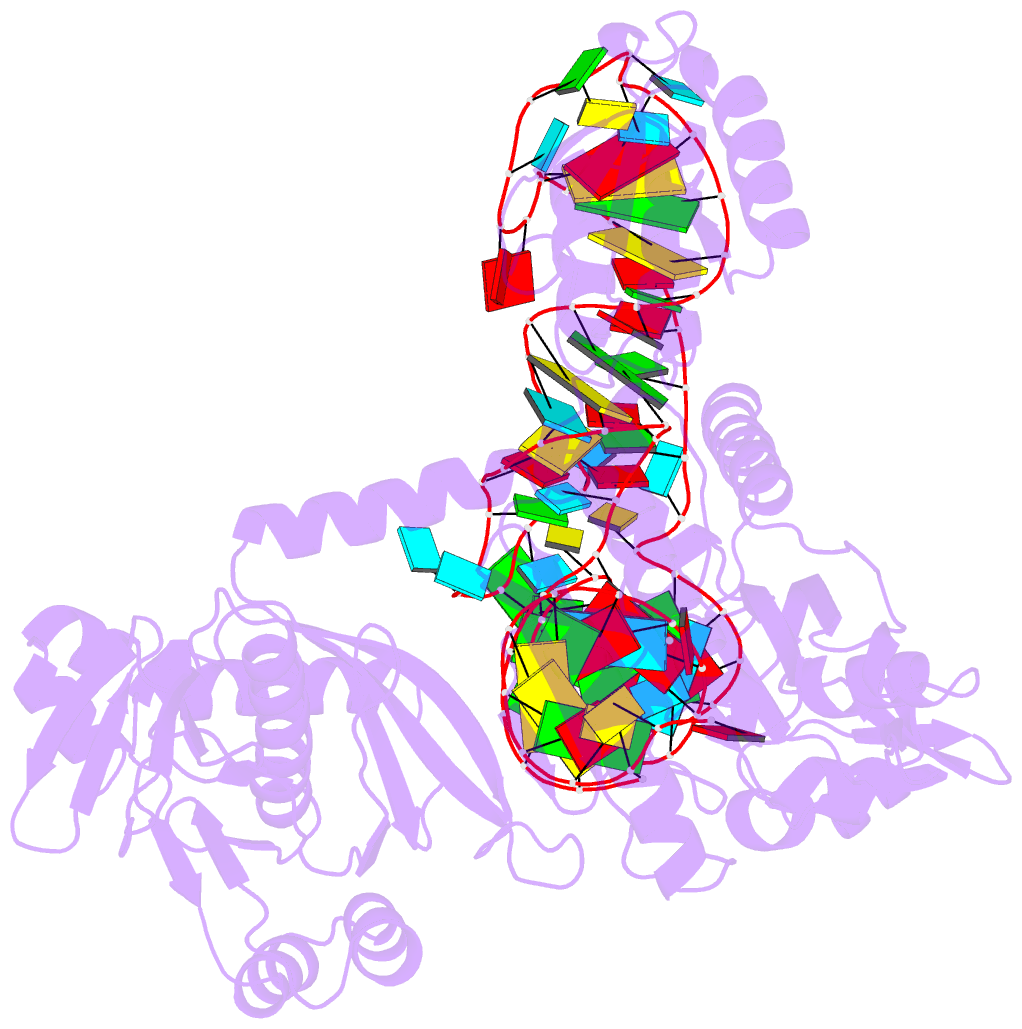
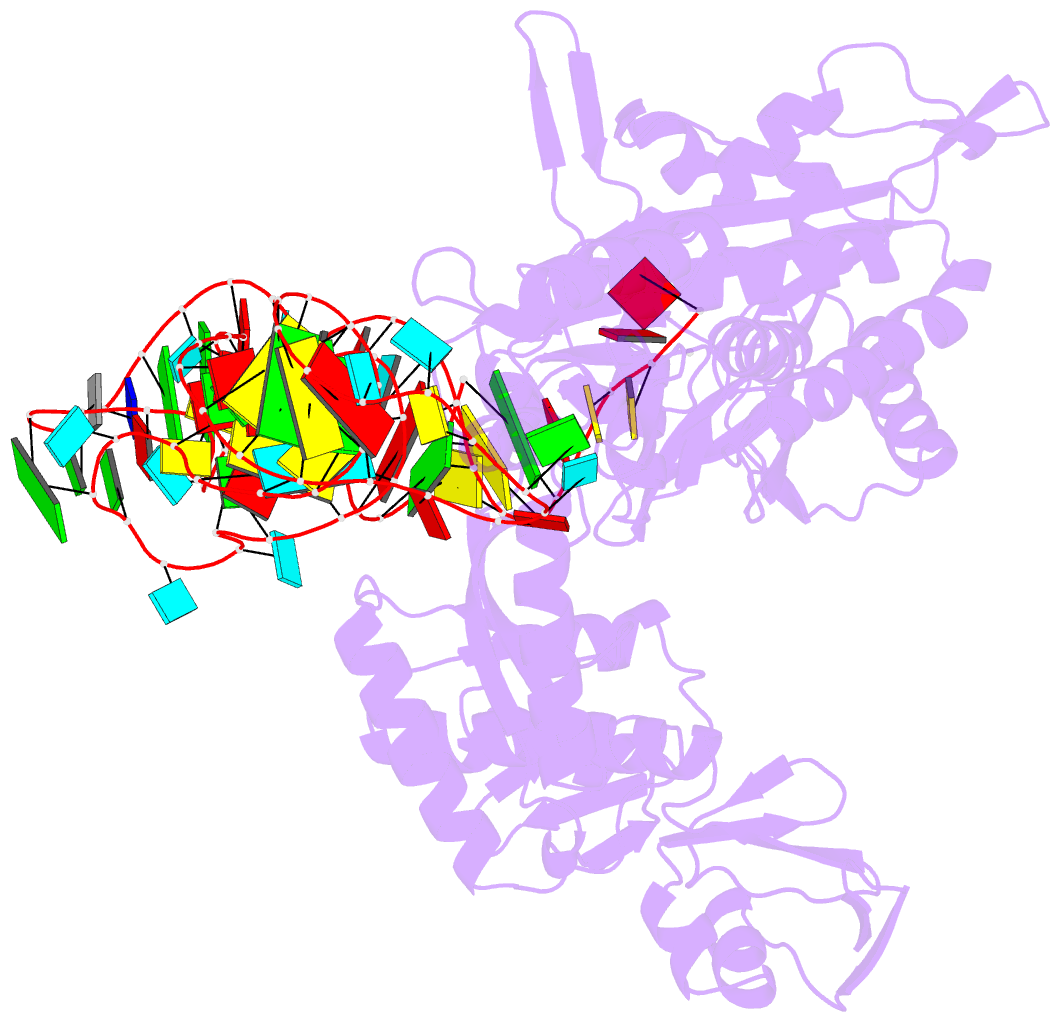
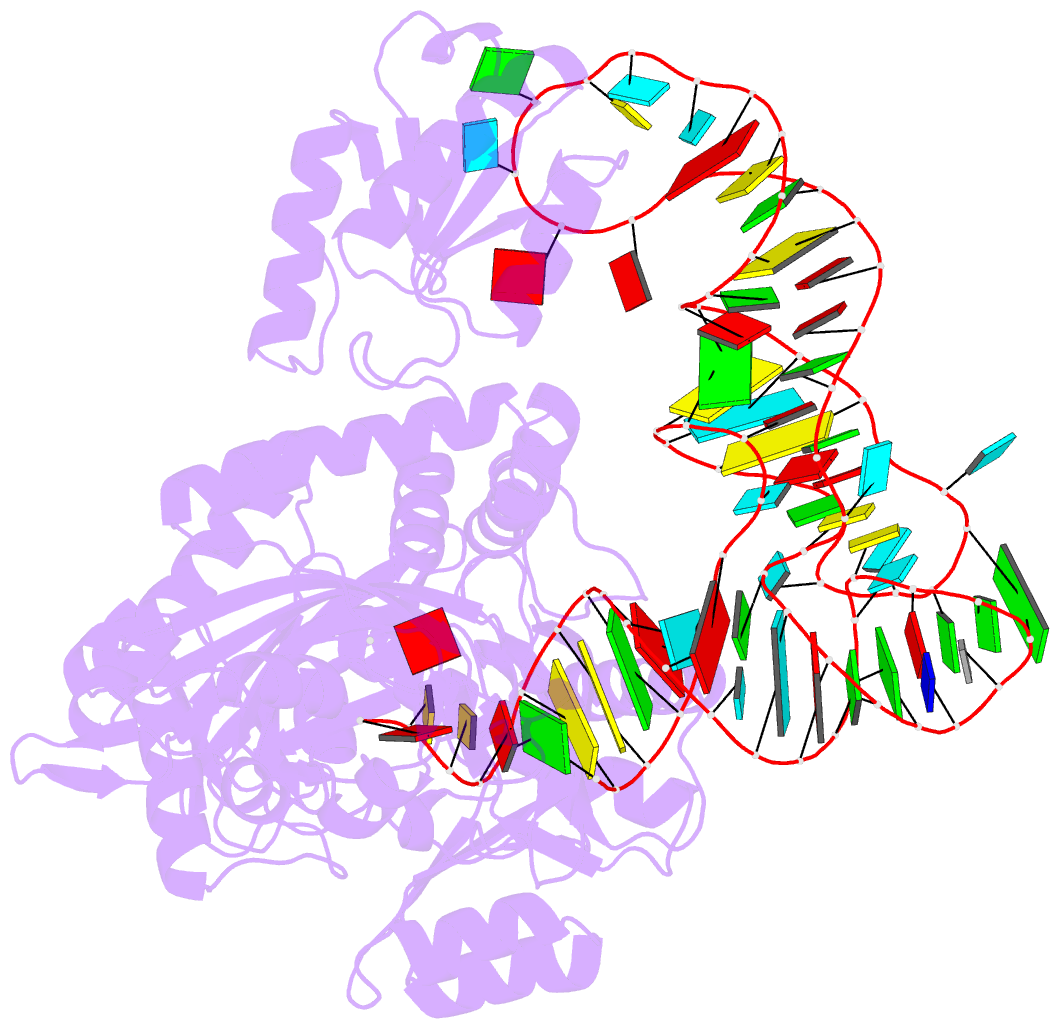
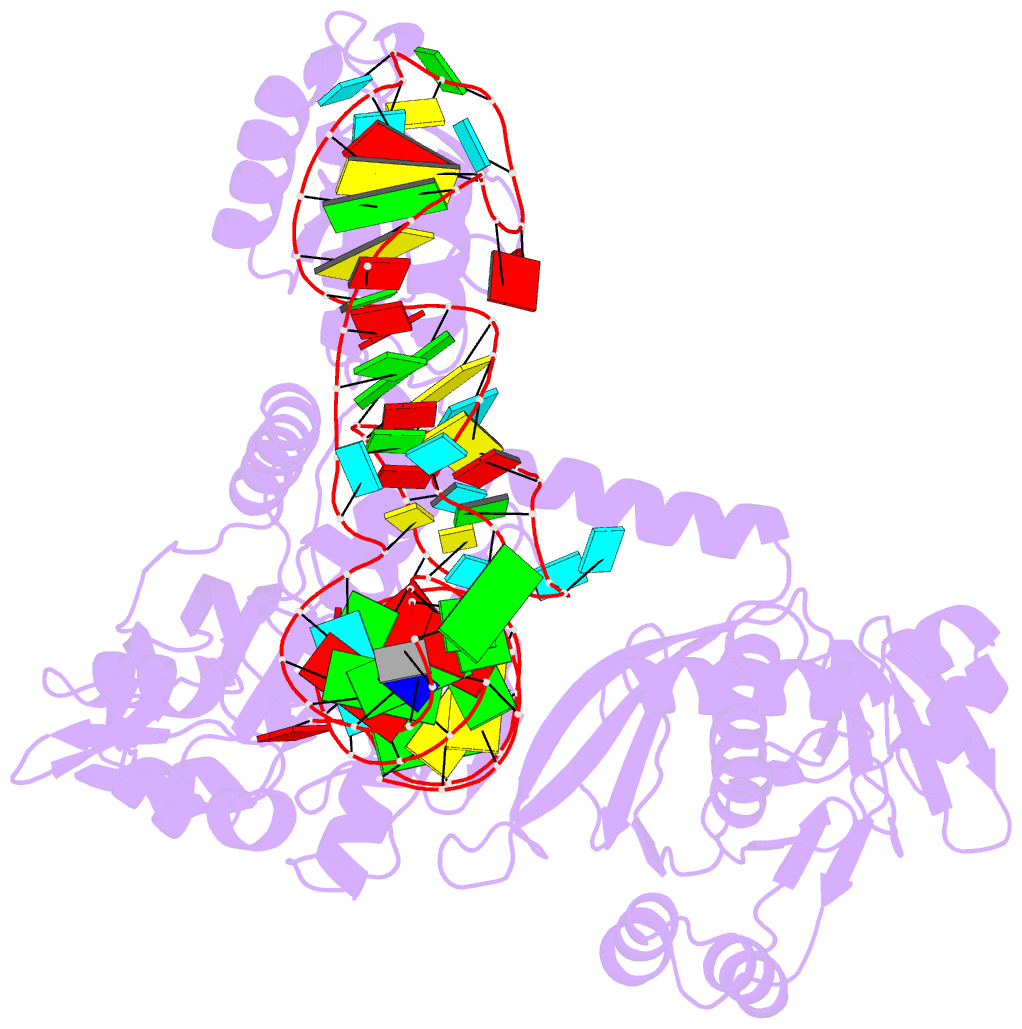
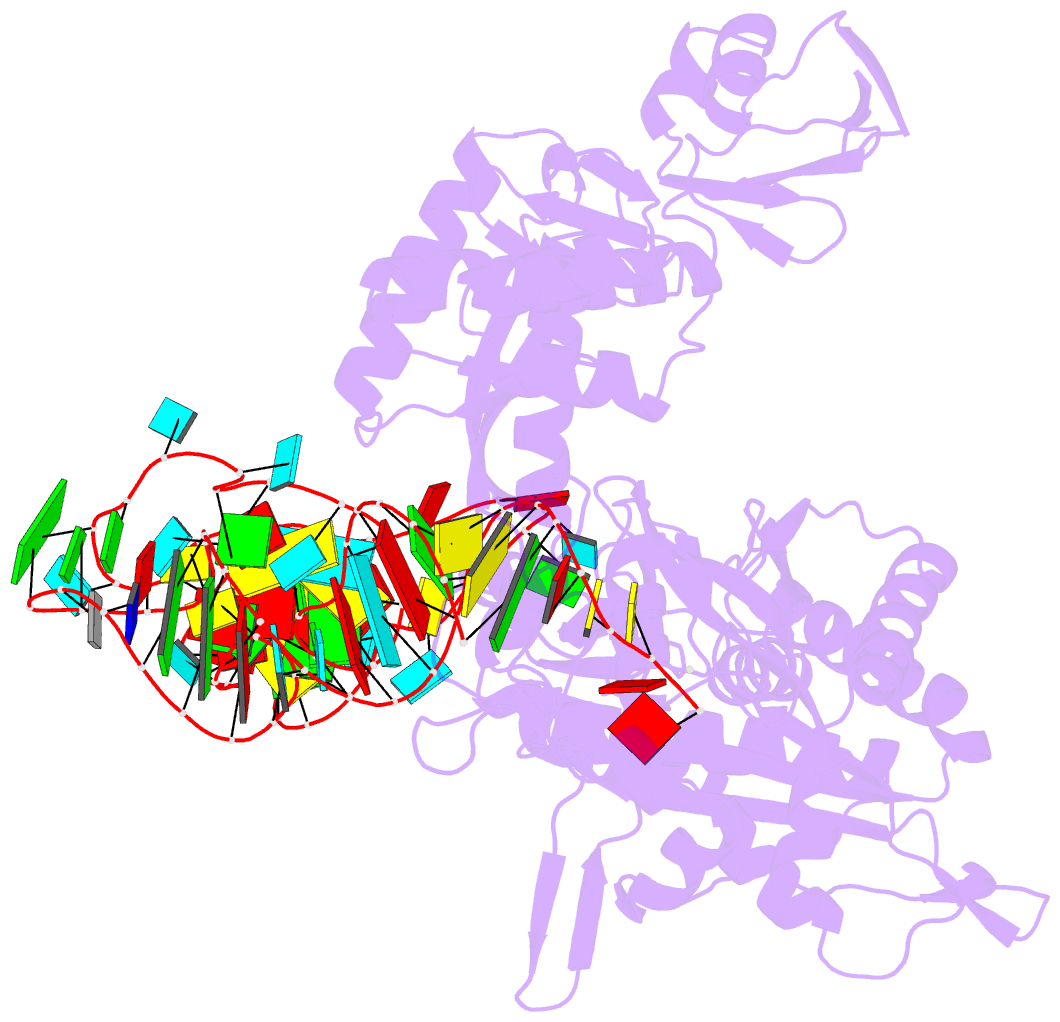
- Structure of e. coli threonyl-trna synthetase complexed with its cognate trna
- Reference
- Sankaranarayanan R, Dock-Bregeon AC, Romby P, Caillet J, Springer M, Rees B, Ehresmann C, Ehresmann B, Moras D (1999): "The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site." Cell(Cambridge,Mass.), 97, 371-381. doi: 10.1016/S0092-8674(00)80746-1.
- Abstract
- E. coli threonyl-tRNA synthetase (ThrRS) is a class II enzyme that represses the translation of its own mRNA. We report the crystal structure at 2.9 A resolution of the complex between tRNA(Thr) and ThrRS, whose structural features reveal novel strategies for providing specificity in tRNA selection. These include an amino-terminal domain containing a novel protein fold that makes minor groove contacts with the tRNA acceptor stem. The enzyme induces a large deformation of the anticodon loop, resulting in an interaction between two adjacent anticodon bases, which accounts for their prominent role in tRNA identity and translational regulation. A zinc ion found in the active site is implicated in amino acid recognition/discrimination.