Summary information and primary citation
- PDB-id
- 1qp9; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.8 Å)
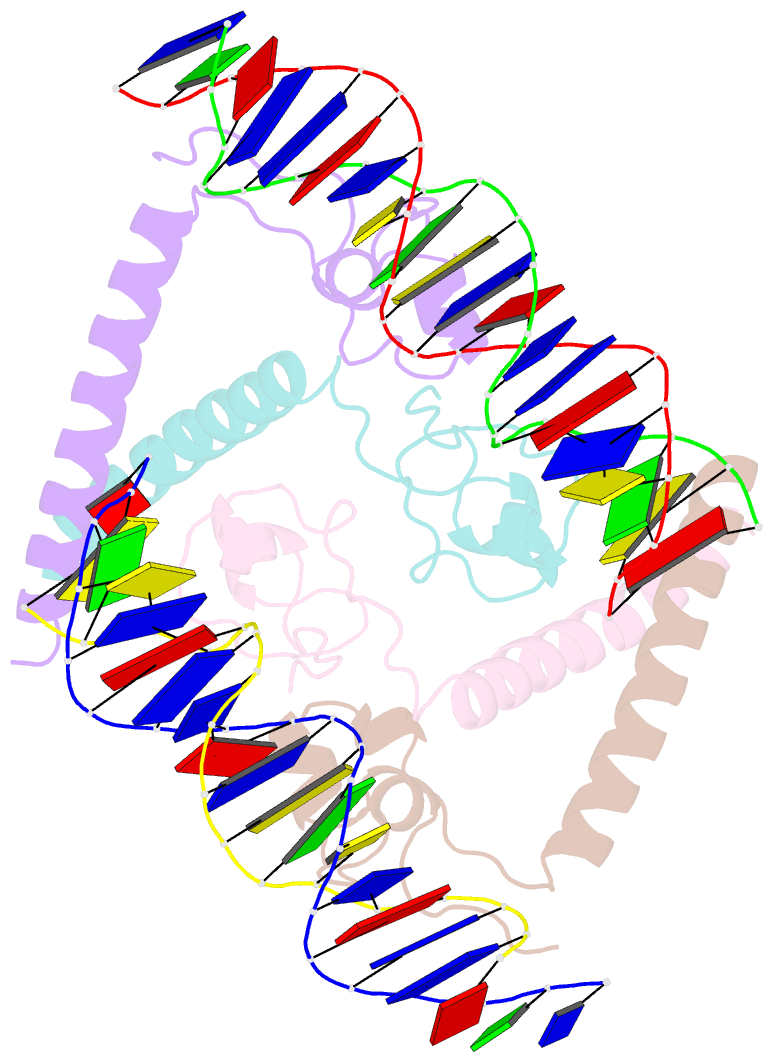
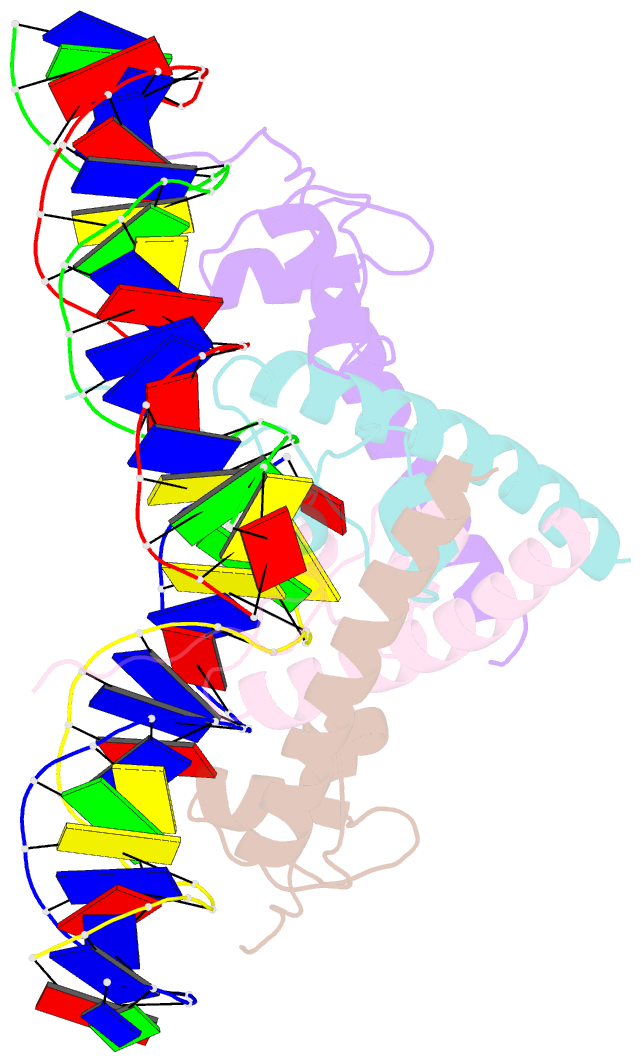
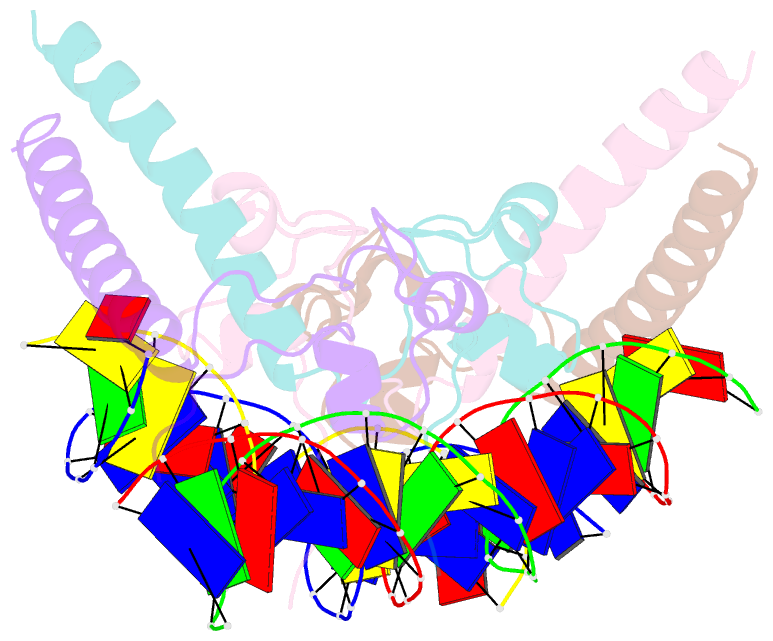
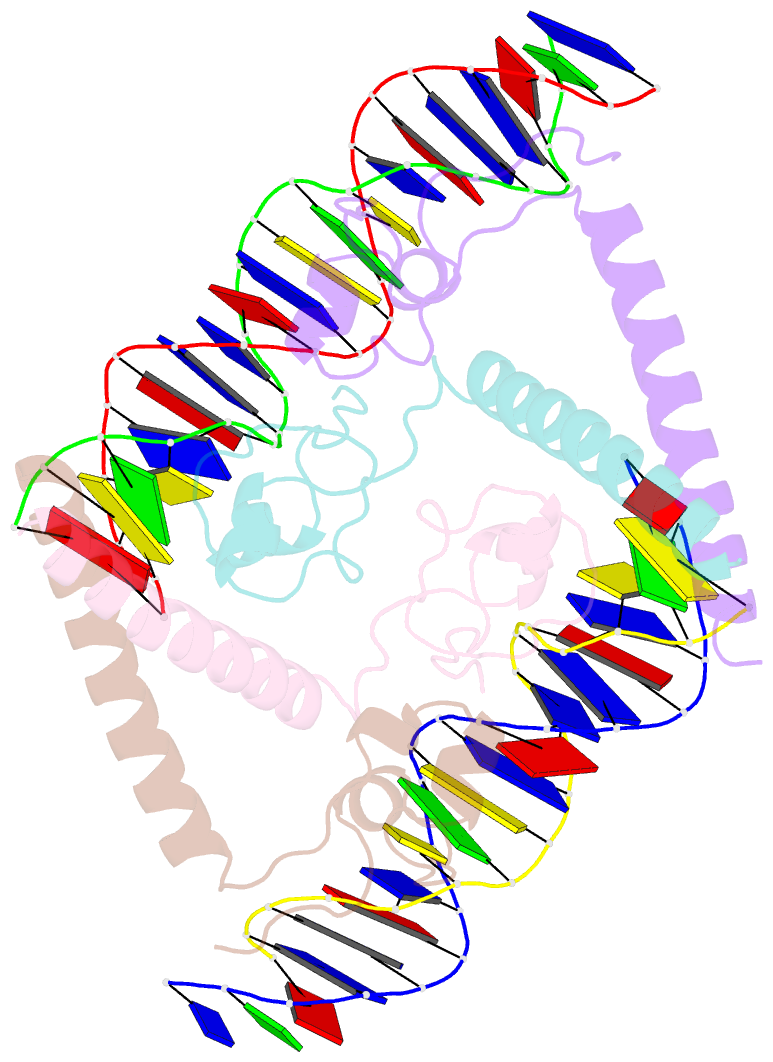
- Summary
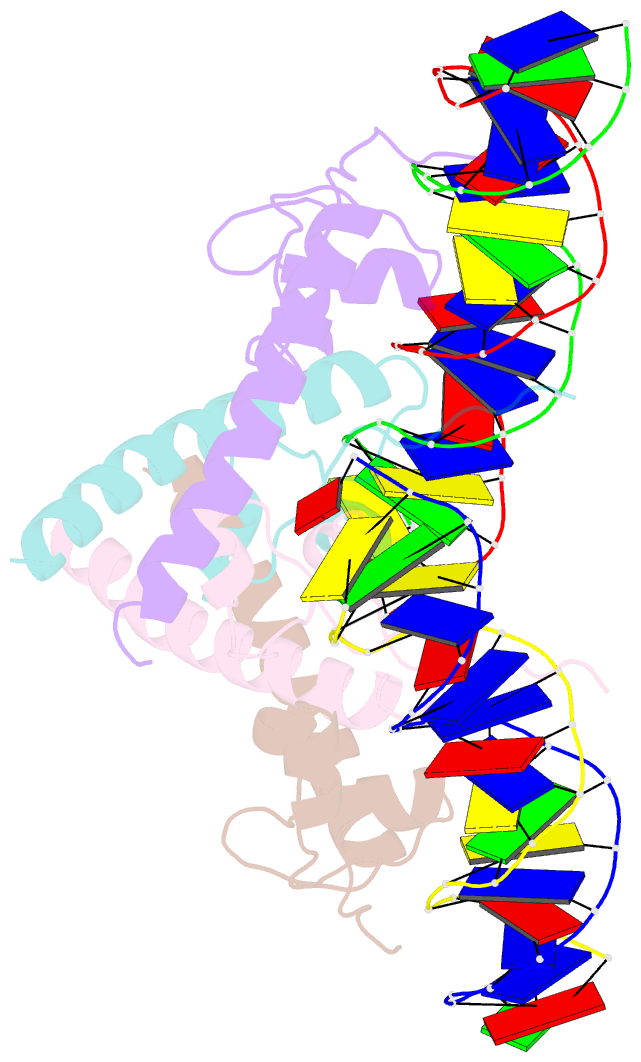
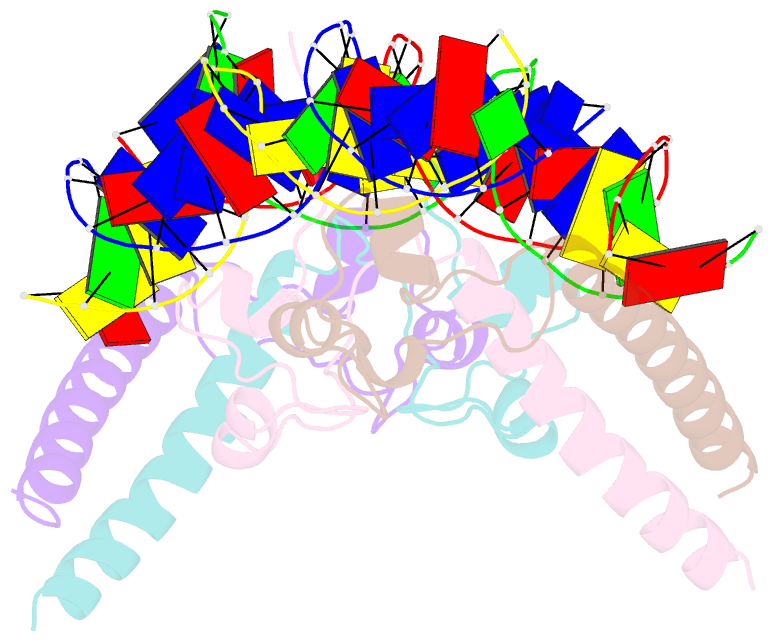
- Structure of hap1-pc7 complexed to the uas of cyc7
- Reference
- Lukens AK, King DA, Marmorstein R (2000): "Structure of HAP1-PC7 bound to DNA: implications for DNA recognition and allosteric effects of DNA-binding on transcriptional activation." Nucleic Acids Res., 28, 3853-3863. doi: 10.1093/nar/28.20.3853.
- Abstract
- HAP1 is a transcription factor in yeast whose DNA-binding domain has been implicated in directly affecting transcriptional activation. Two separate mutations in the DNA-binding domain, S63G (HAP1-PC7) and S63R (HAP1-18), retain wild-type binding affinity. However, HAP1-PC7 is transcriptionally silent while HAP1-18 shows highly elevated levels of transcription. We have determined the X-ray crystal structure of the DNA-binding domain of HAP1-PC7 bound to its DNA target, UAS(CYC7), and compared it to the previously solved HAP1-wt and HAP1-18 complexes to UAS(CYC7). Additionally, we have quantitatively compared the DNA-binding affinity and specificity of the HAP1-PC7, HAP1-18 and HAP1-wt DNA-binding domains. We show that, although the DNA-binding domains of these three proteins bind UAS(CYC7) with comparable affinity and specificity, the protein-DNA interactions are dramatically different between the three complexes. Conserved protein-DNA interactions are largely restricted to an internal DNA sequence that excludes one of the two conserved DNA half-sites of UAS(CYC7) suggesting a mode of recognition distinct from other HAP1 family members. Alternative protein-DNA interactions result in divergent DNA configurations between the three complexes. These results suggest that the differential transcriptional activities of the HAP1, HAP1-18 and HAP1-PC7 proteins are due, at least in part, to alternative protein-DNA contacts, and implies that HAP1-DNA interactions have direct allosteric effects on transcriptional activation.