Summary information and primary citation
- PDB-id
- 1qpi; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.5 Å)
- Summary
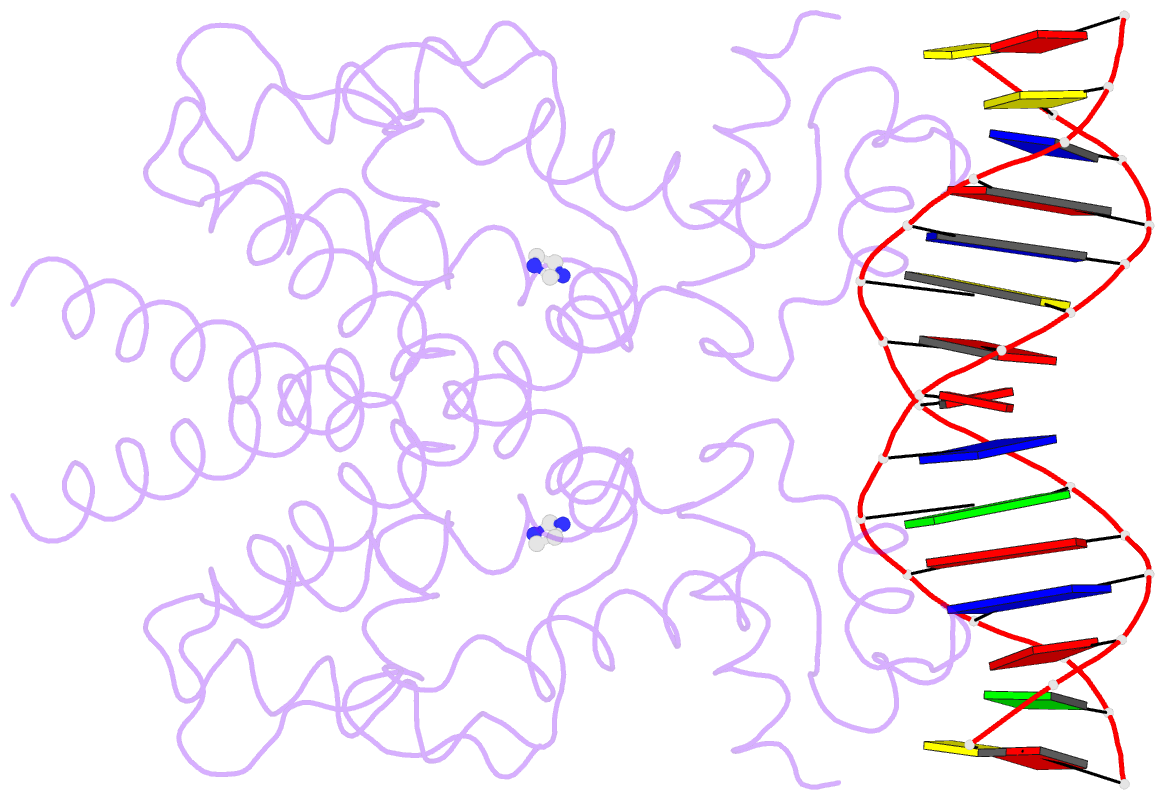
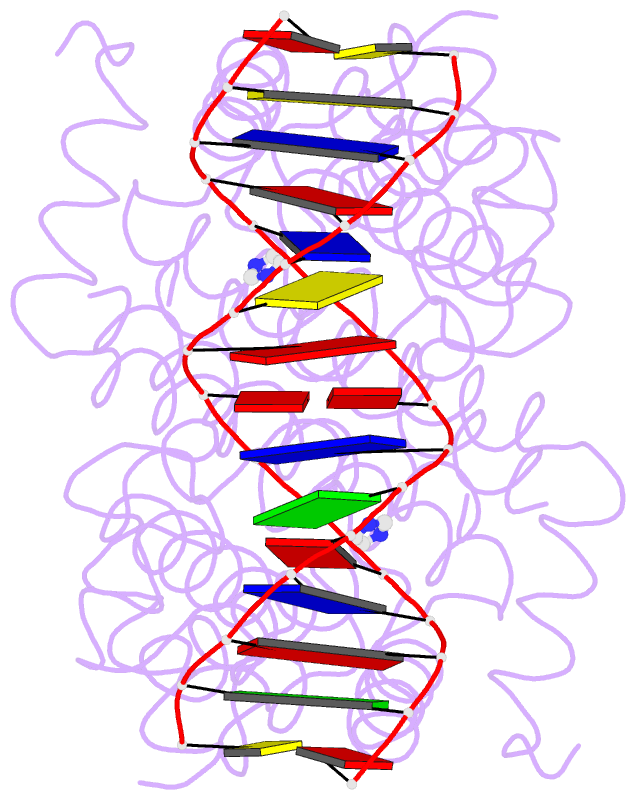
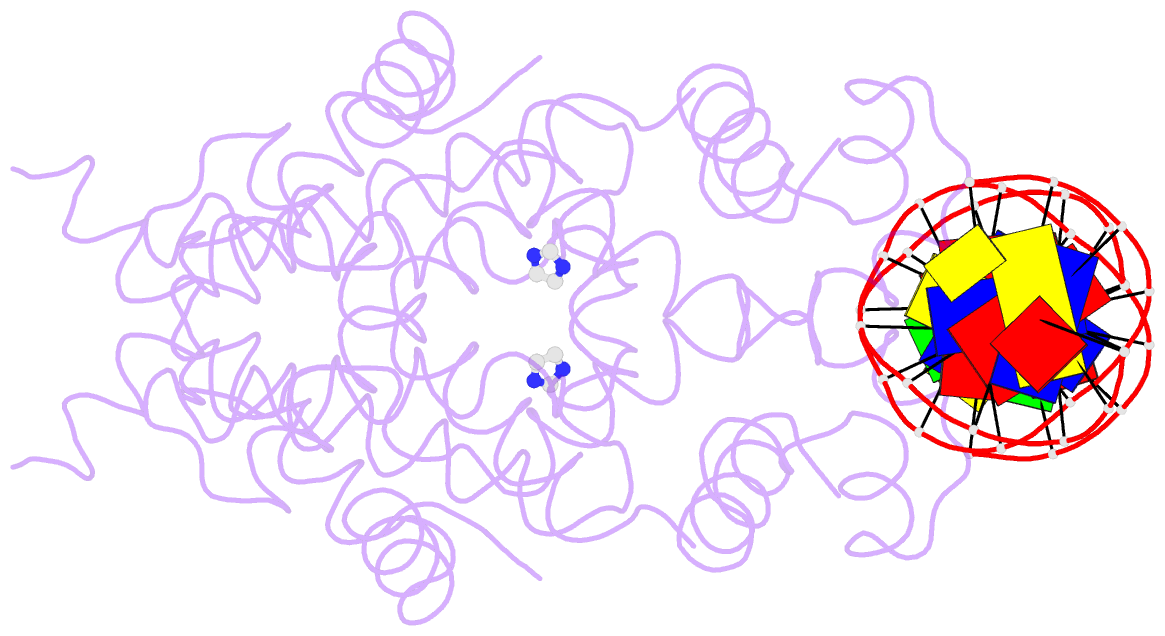
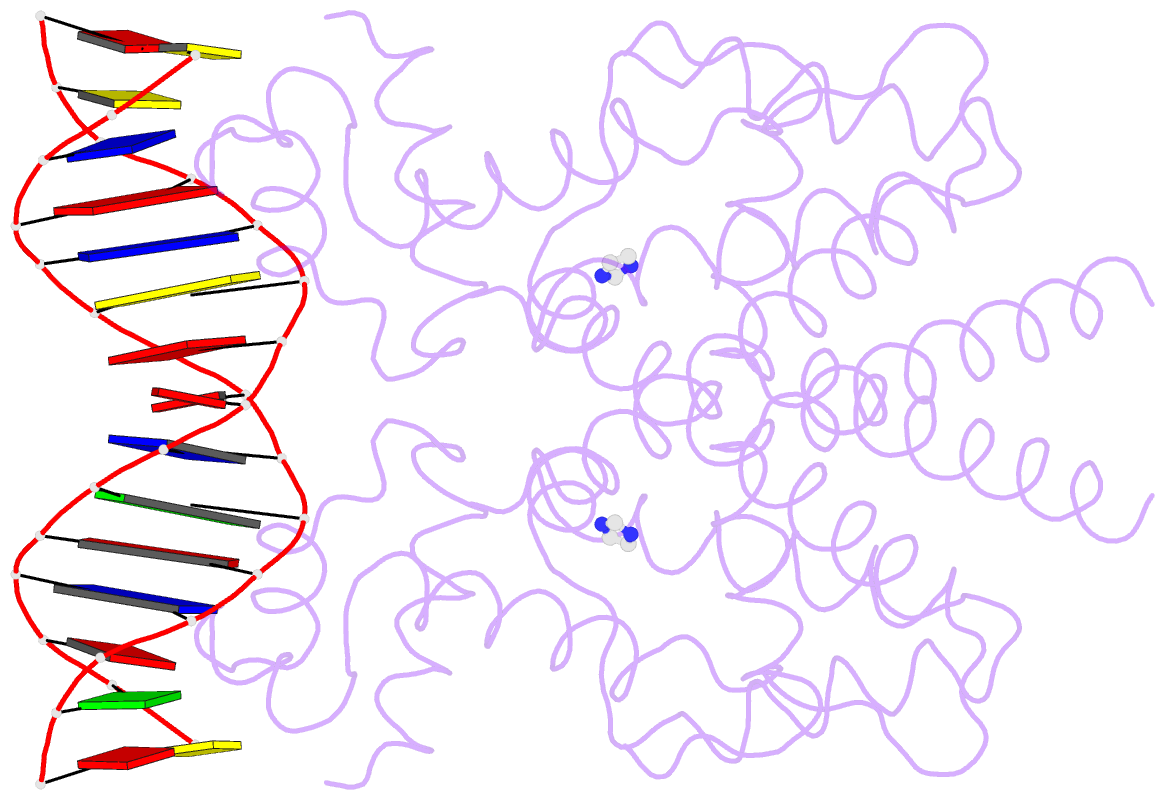
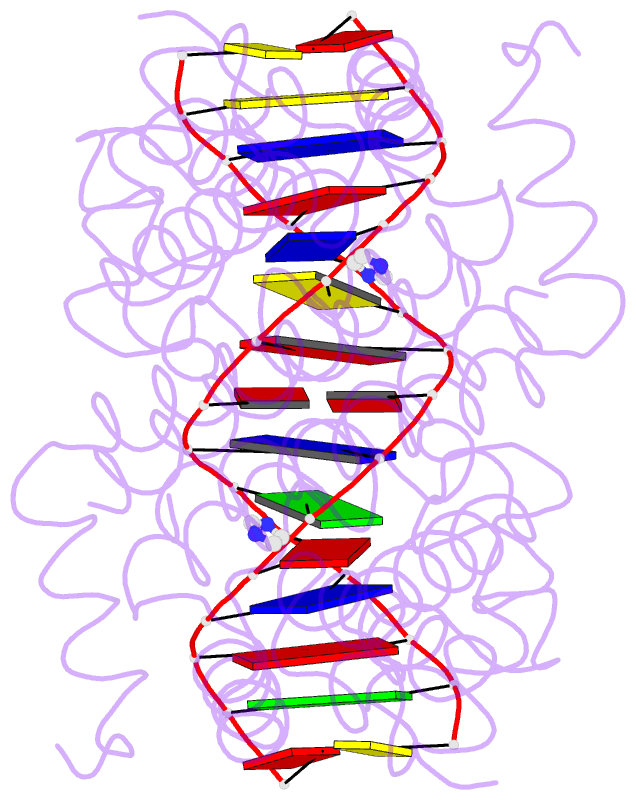
- Crystal structure of tetracycline repressor-operator complex
- Reference
- Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W (2000): "Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system." Nat.Struct.Biol., 7, 215-219. doi: 10.1038/73324.
- Abstract
- The tetracycline repressor (TetR) regulates the most abundant resistance mechanism against the antibiotic tetracycline in grain-negative bacteria. The TetR protein and its mutants are commonly used as control elements to regulate gene expression in higher eukaryotes. We present the crystal structure of the TetR homodimer in complex with its palindromic DNA operator at 2.5 A resolution. Comparison to the structure of TetR in complex with the inducer tetracycline-Mg2+ allows the mechanism of induction to be deduced. Inducer binding in the repressor core initiates conformational changes starting with C-terminal unwinding and shifting of the short helix a6 in each monomer. This forces a pendulum-like motion of helix a4, which increases the separation of the attached DNA binding domains by 3 A, abolishing the affinity of TetR for its operator DNA.