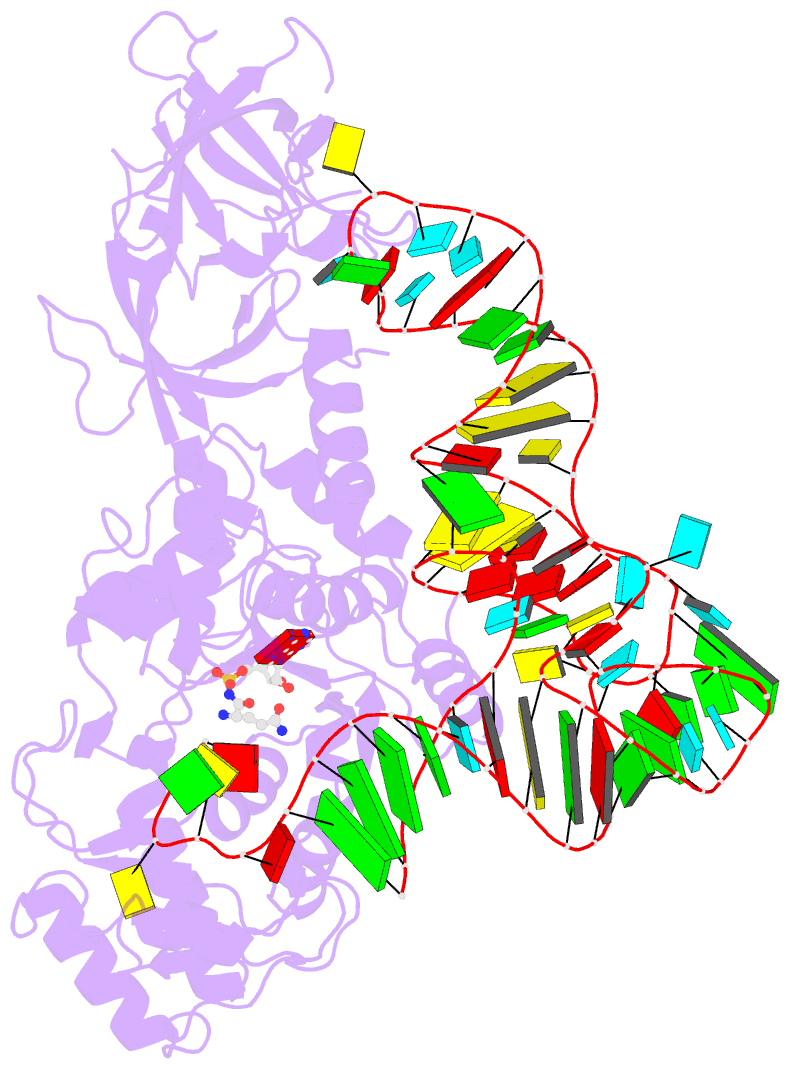
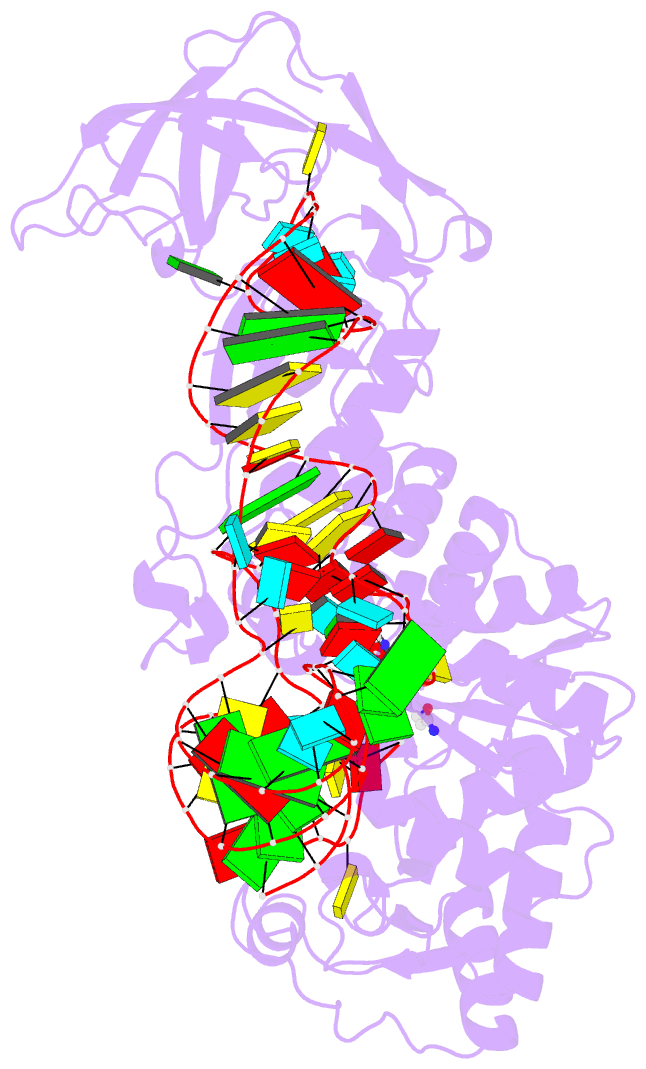
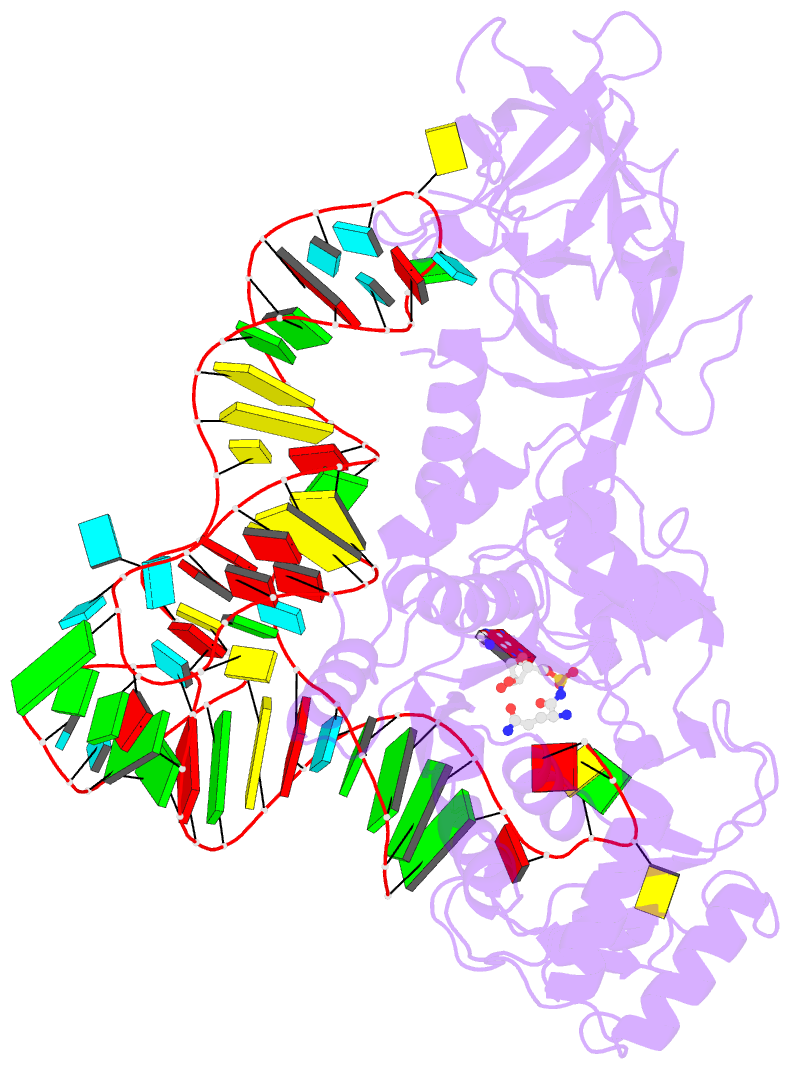
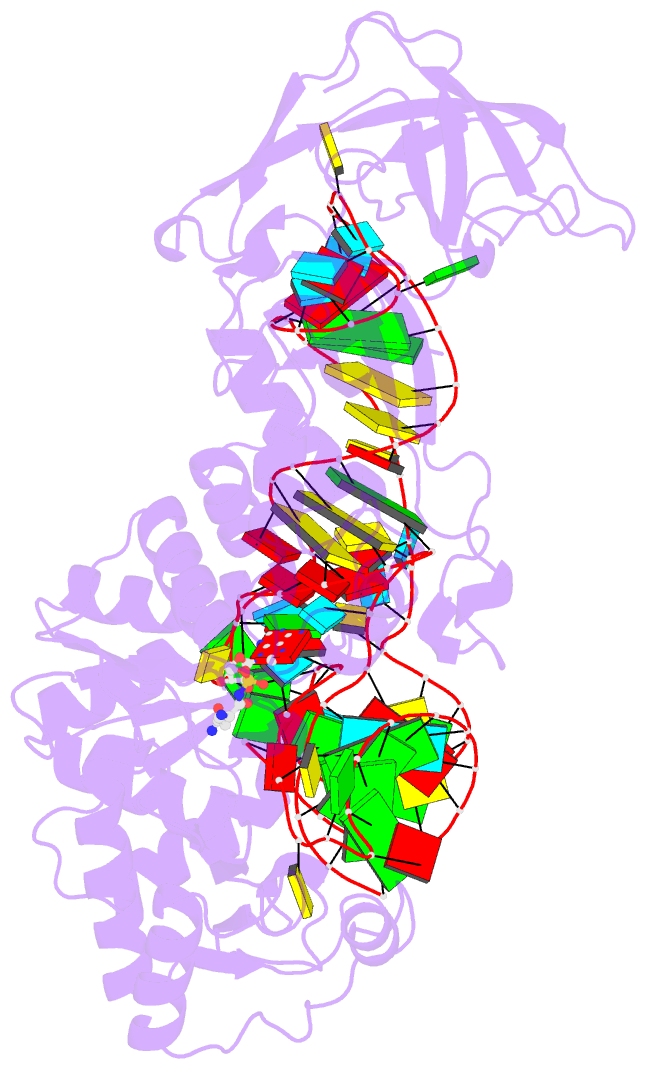
Summary information and primary citation
- PDB-id
- 1qtq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ligase-RNA
- Method
- X-ray (2.25 Å)
- Summary
- Glutaminyl-trna synthetase complexed with trna and an amino acid analog
- Reference
- Rath VL, Silvian LF, Beijer B, Sproat BS, Steitz TA (1998): "How glutaminyl-tRNA synthetase selects glutamine." Structure, 6, 439-449. doi: 10.1016/S0969-2126(98)00046-X.
- Abstract
- Background: Aminoacyl-tRNA synthetases covalently link a specific amino acid to the correct tRNA. The fidelity of this reaction is essential for accurate protein synthesis. Each synthetase has a specific molecular mechanism to distinguish the correct pair of substrates from the pool of amino acids and isologous tRNA molecules. In the case of glutaminyl-tRNA synthetase (GlnRS) the prior binding of tRNA is required for activation of glutamine by ATP. A complete understanding of amino acid specificity in GlnRS requires the determination of the structure of the synthetase with both tRNA and substrates bound.
Results: A stable glutaminly-adenylate analog, which inhibits GlnRS with a Ki of 1.32 microM, was synthesized and cocrystallized with GlnRS and tRNA2Gln. The crystal structure of this ternary complex has been refined at 2.4 A resolution and shows the interactions made between glutamine and its binding site.
Conclusions: To select against glutamic acid or glutamate, both hydrogen atoms of the nitrogen of the glutamine sidechain are recognized. The hydroxyl group of Tyr211 and a water molecule are responsible for this recognition; both are obligate hydrogen-bond acceptors due to a network of interacting sidechains and water molecules. The prior binding of tRNAGln that is required for amino acid activation may result from the terminal nucleotide, A76, packing against and orienting Tyr211, which forms part of the amino acid binding site.