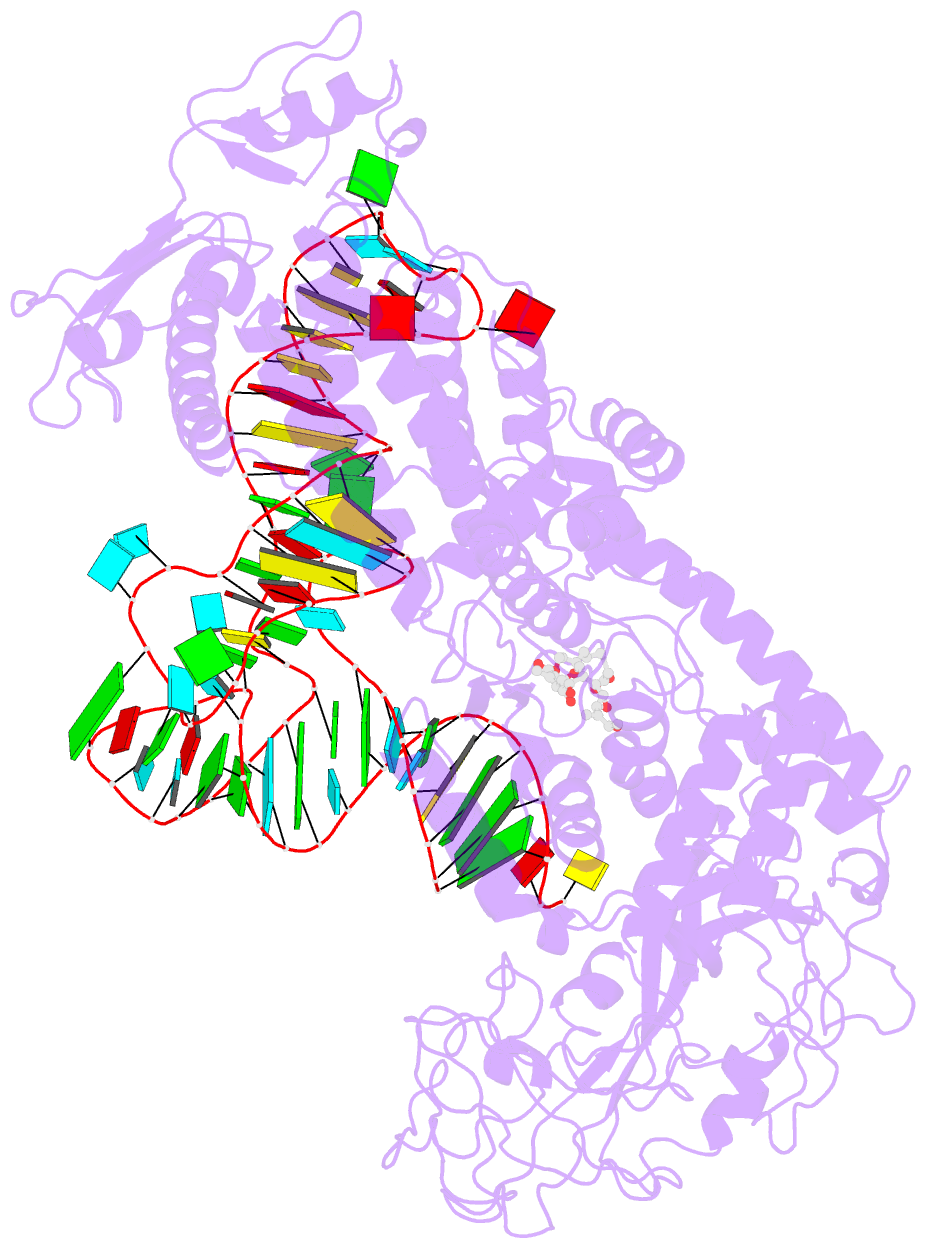
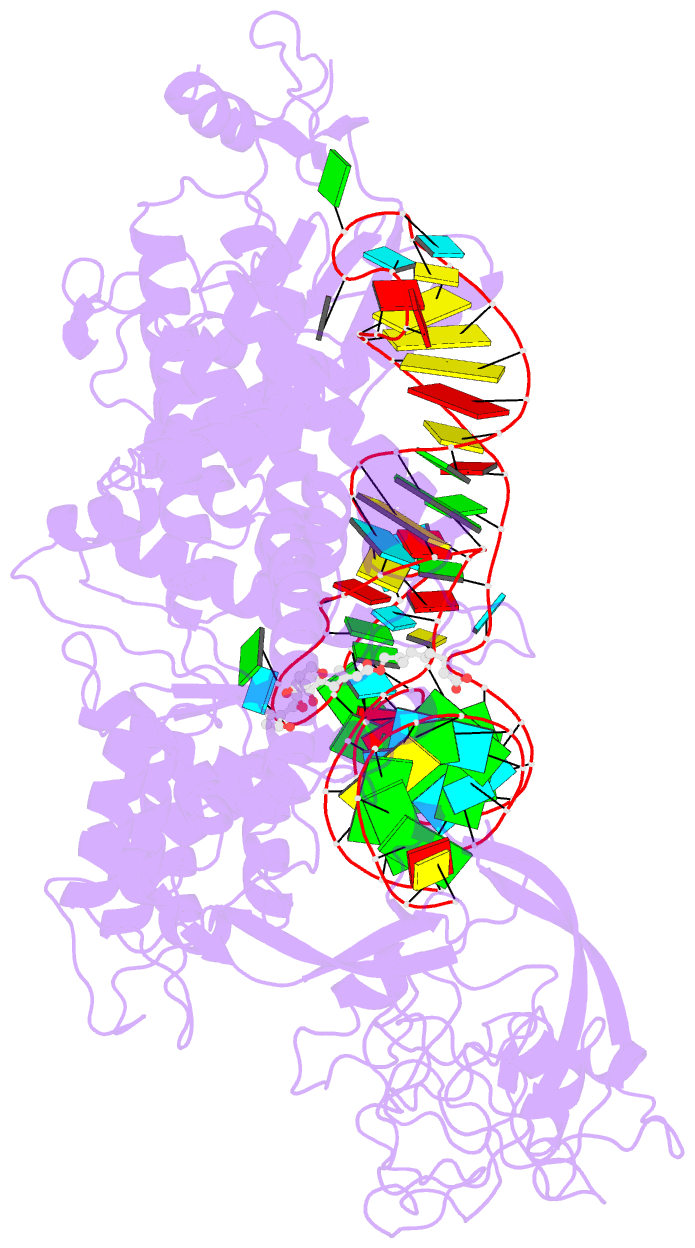
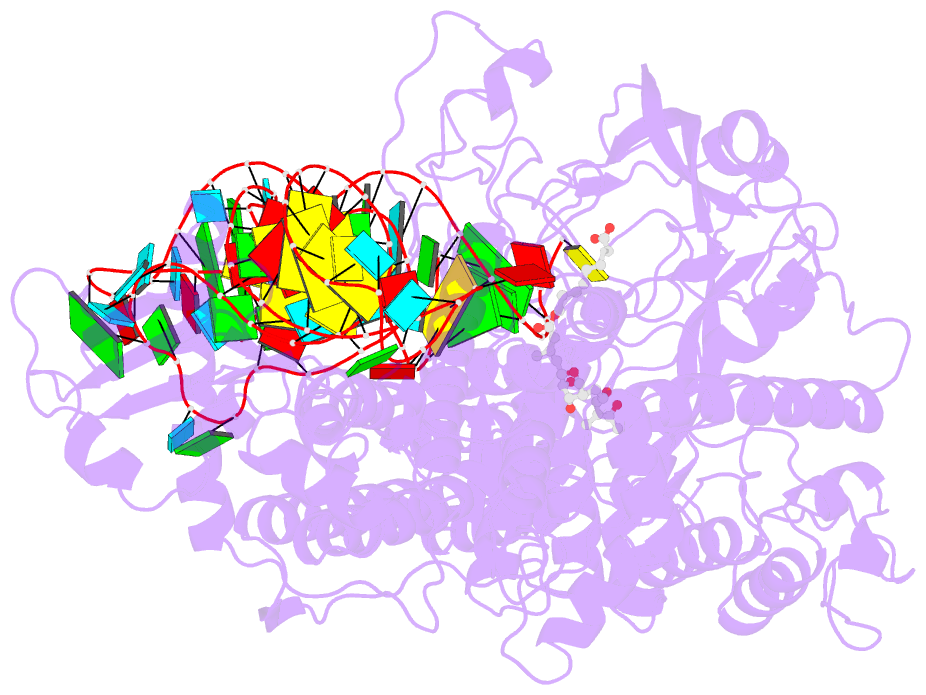
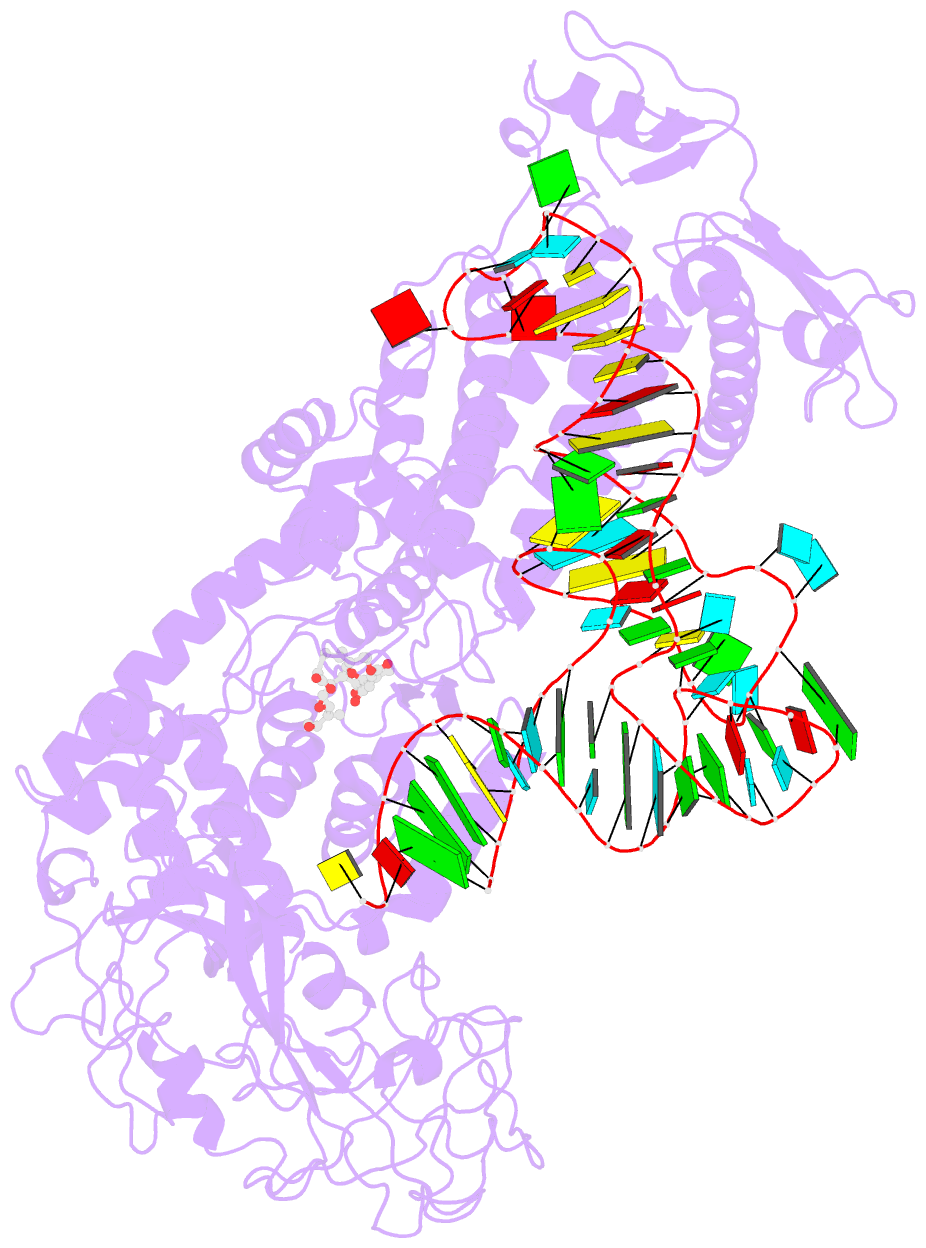
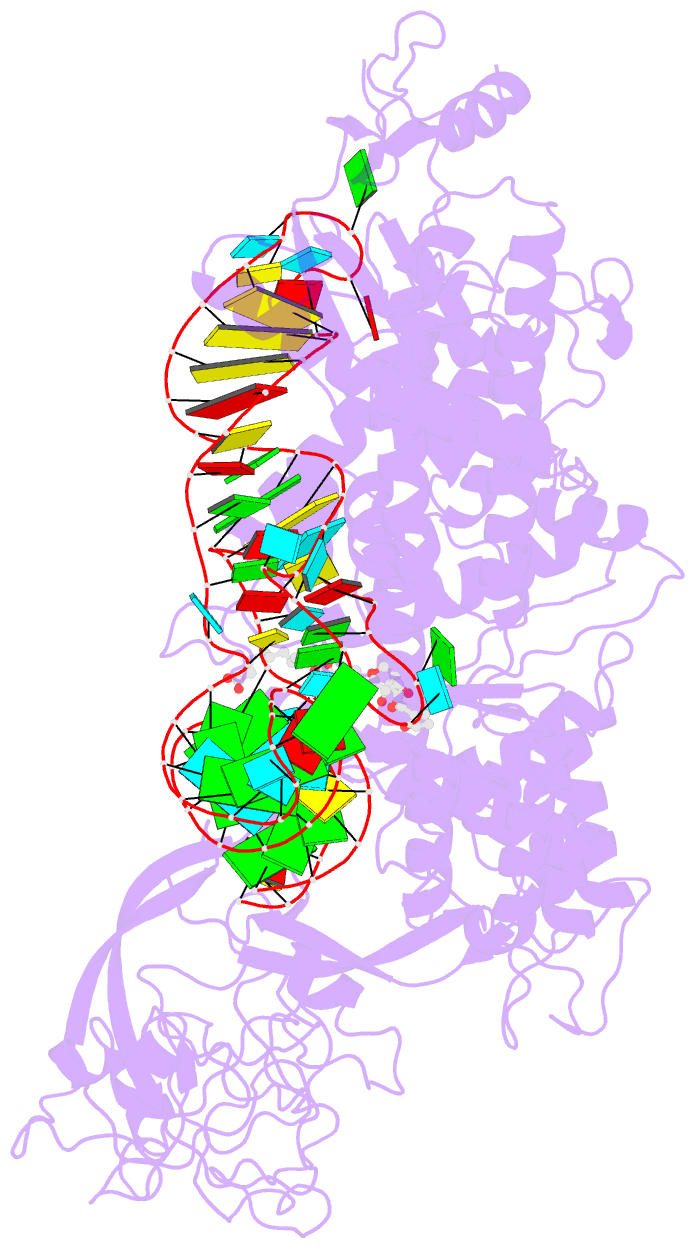
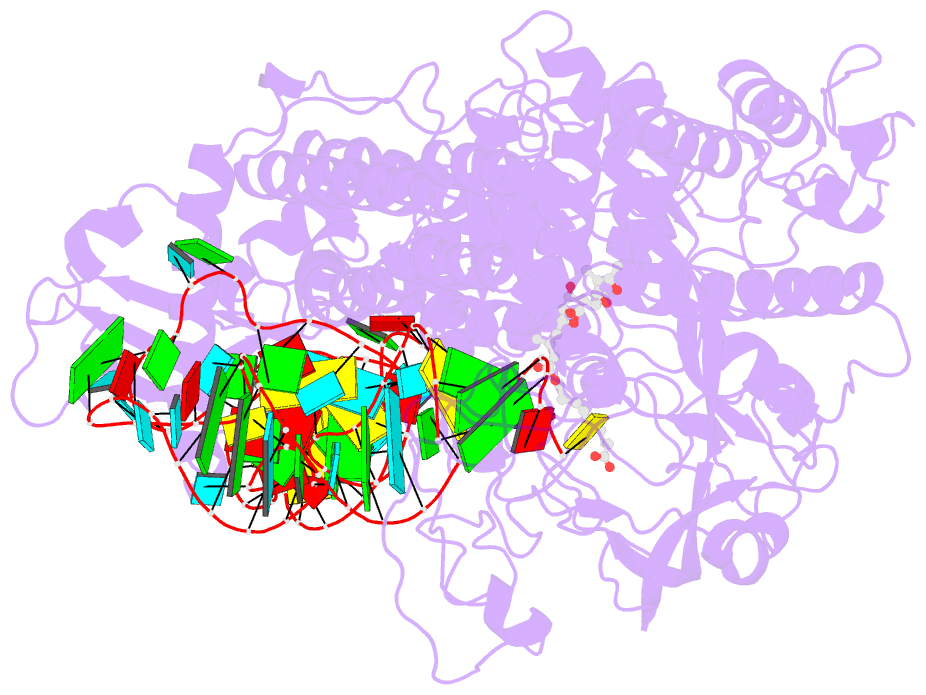
Summary information and primary citation
- PDB-id
- 1qu2; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ligase-RNA
- Method
- X-ray (2.2 Å)
- Summary
- Insights into editing from an ile-trna synthetase structure with trna(ile) and mupirocin
- Reference
- Silvian LF, Wang J, Steitz TA (1999): "Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin." Science, 285, 1074-1077. doi: 10.1126/science.285.5430.1074.
- Abstract
- Isoleucyl-transfer RNA (tRNA) synthetase (IleRS) joins Ile to tRNA(Ile) at its synthetic active site and hydrolyzes incorrectly acylated amino acids at its editing active site. The 2.2 angstrom resolution crystal structure of Staphylococcus aureus IleRS complexed with tRNA(Ile) and Mupirocin shows the acceptor strand of the tRNA(Ile) in the continuously stacked, A-form conformation with the 3' terminal nucleotide in the editing active site. To position the 3' terminus in the synthetic active site, the acceptor strand must adopt the hairpinned conformation seen in tRNA(Gln) complexed with its synthetase. The amino acid editing activity of the IleRS may result from the incorrect products shuttling between the synthetic and editing active sites, which is reminiscent of the editing mechanism of DNA polymerases.