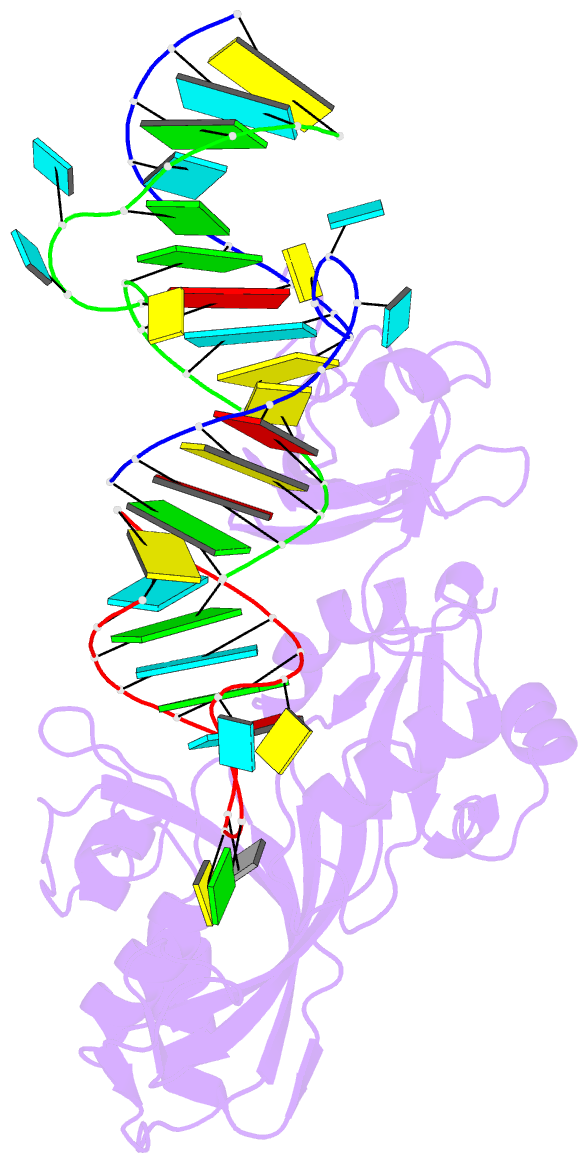
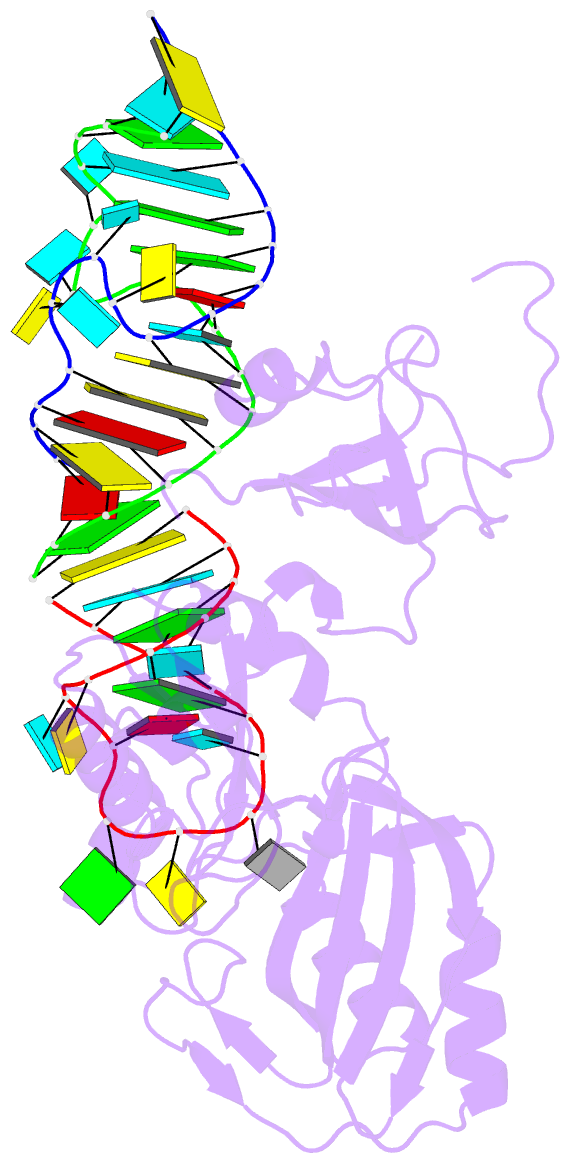
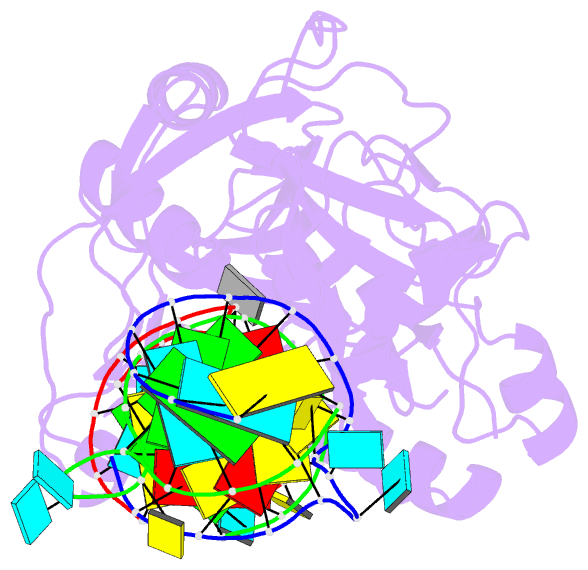
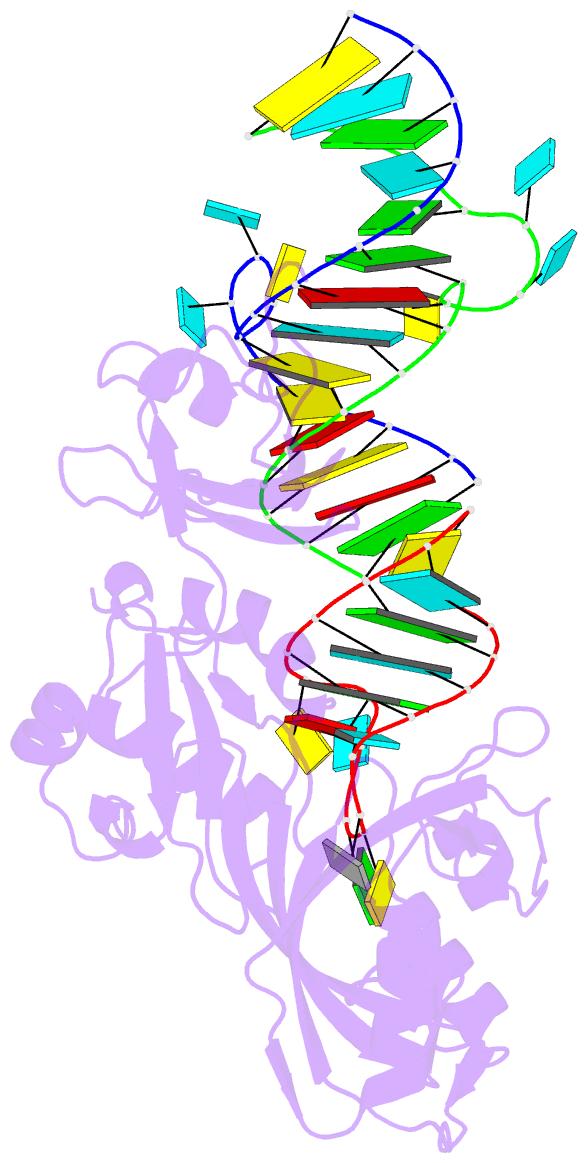
Summary information and primary citation
- PDB-id
- 1r3e; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- lyase-RNA
- Method
- X-ray (2.1 Å)
- Summary
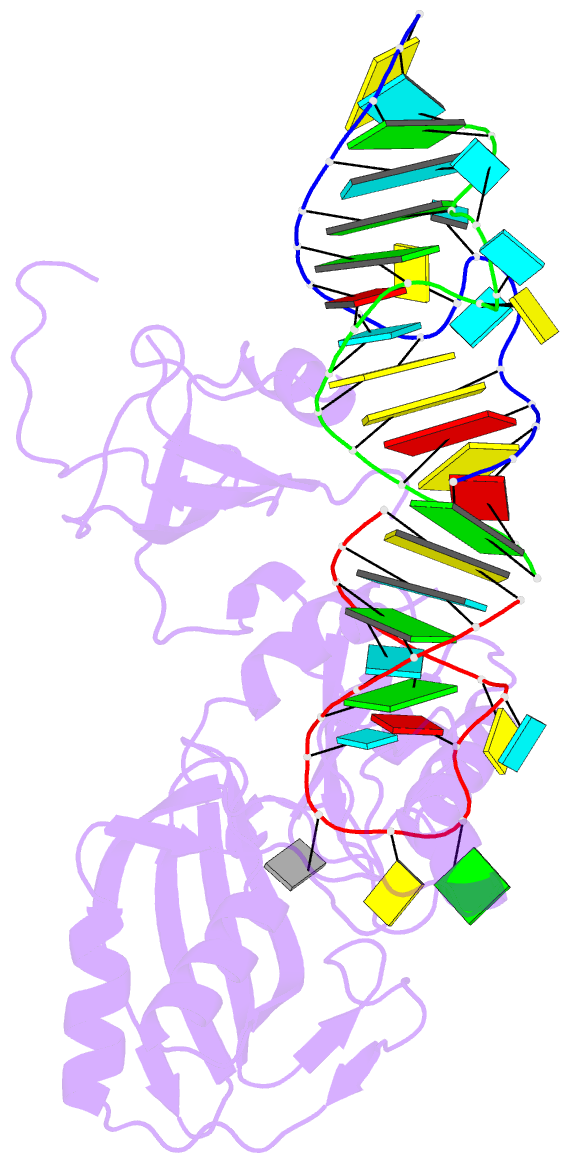
- Crystal structure of trna pseudouridine synthase trub and its RNA complex: RNA-protein recognition through a combination of rigid docking and induced fit
- Reference
- Pan H, Agarwalla S, Moustakas DT, Finer-Moore J, Stroud RM (2003): "Crystal Structure of tRNA Pseudouridine Synthase TruB and Its RNA Complex: RNA Recognition Through a Combination of Rigid Docking and Induced Fit." Proc.Natl.Acad.Sci.USA, 100, 12648-12653. doi: 10.1073/pnas.2135585100.
- Abstract
- RNA pseudouridine synthase, TruB, catalyzes pseudouridine formation at U55 in tRNA. This posttranscriptional modification is almost universally conserved and occurs in the T arm of most tRNAs. We determined the crystal structure of Escherichia coli TruB apo enzyme, as well as the structure of Thermotoga maritima TruB in complex with RNA. Comparison of the RNA-free and -bound forms of TruB reveals that this enzyme undergoes significant conformational changes on binding to its substrate. These conformational changes include the ordering of the "thumb loop," which binds right into the RNA hairpin loop, and a 10 degree hinge movement of the C-terminal domain. Along with the result of docking experiments performed on apo TruB, we conclude that TruB recognizes its RNA substrate through a combination of rigid docking and induced fit, with TruB first rigidly binding to its target and then maximizing the interaction by induced fit.