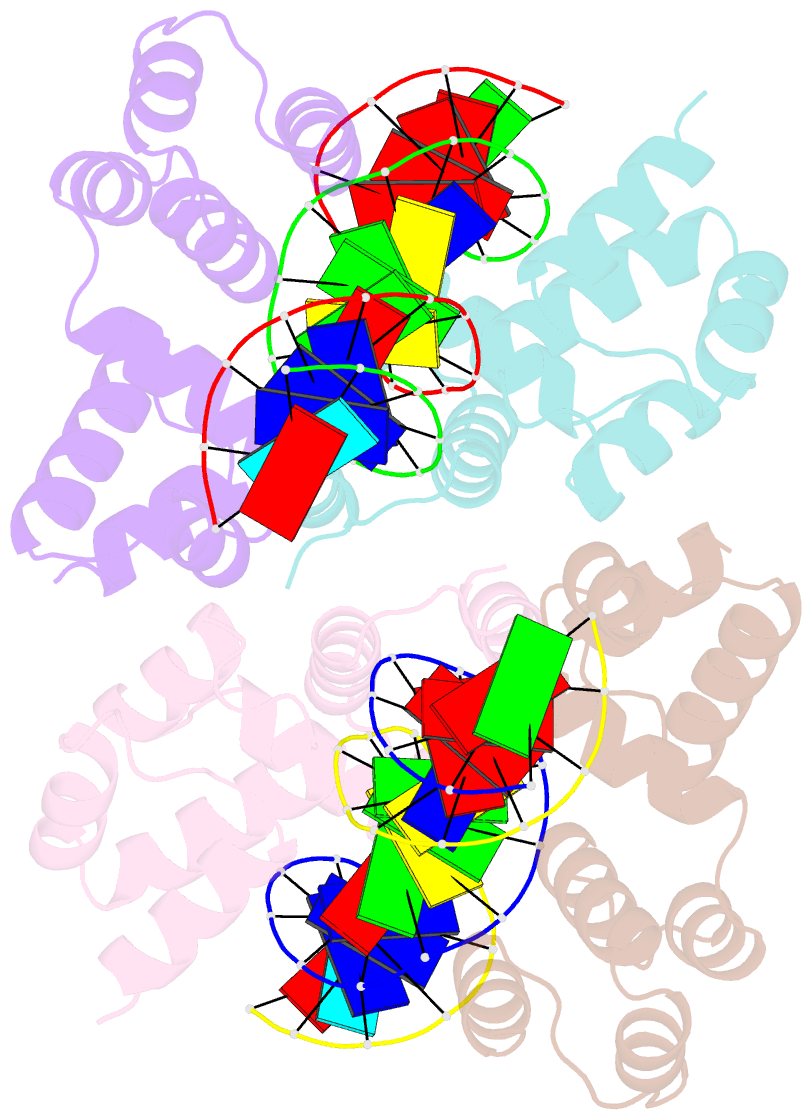
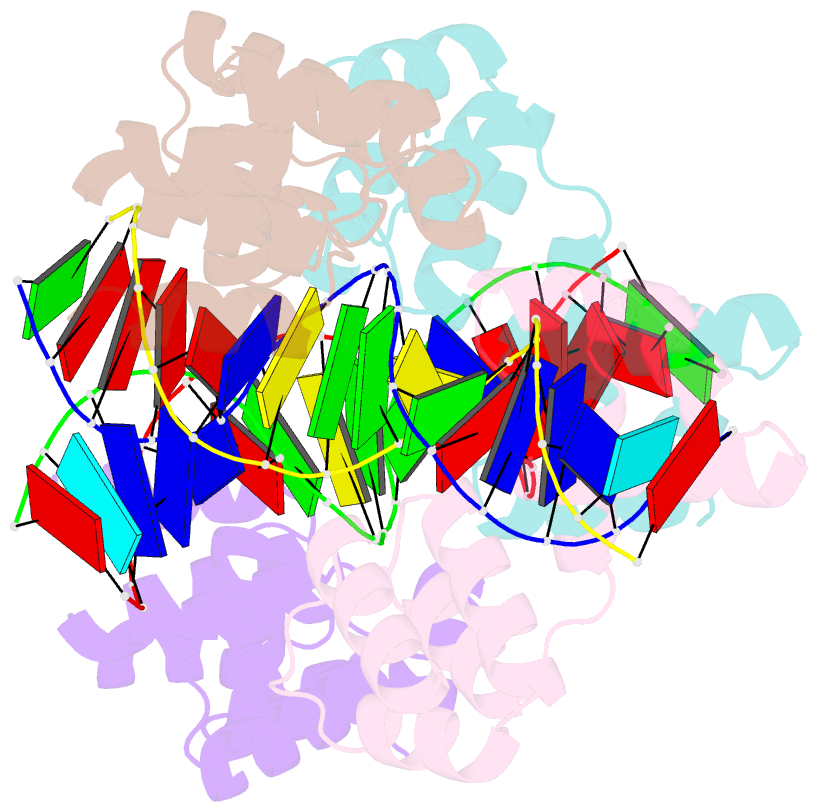
Summary information and primary citation
- PDB-id
- 1r71; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.2 Å)
- Summary
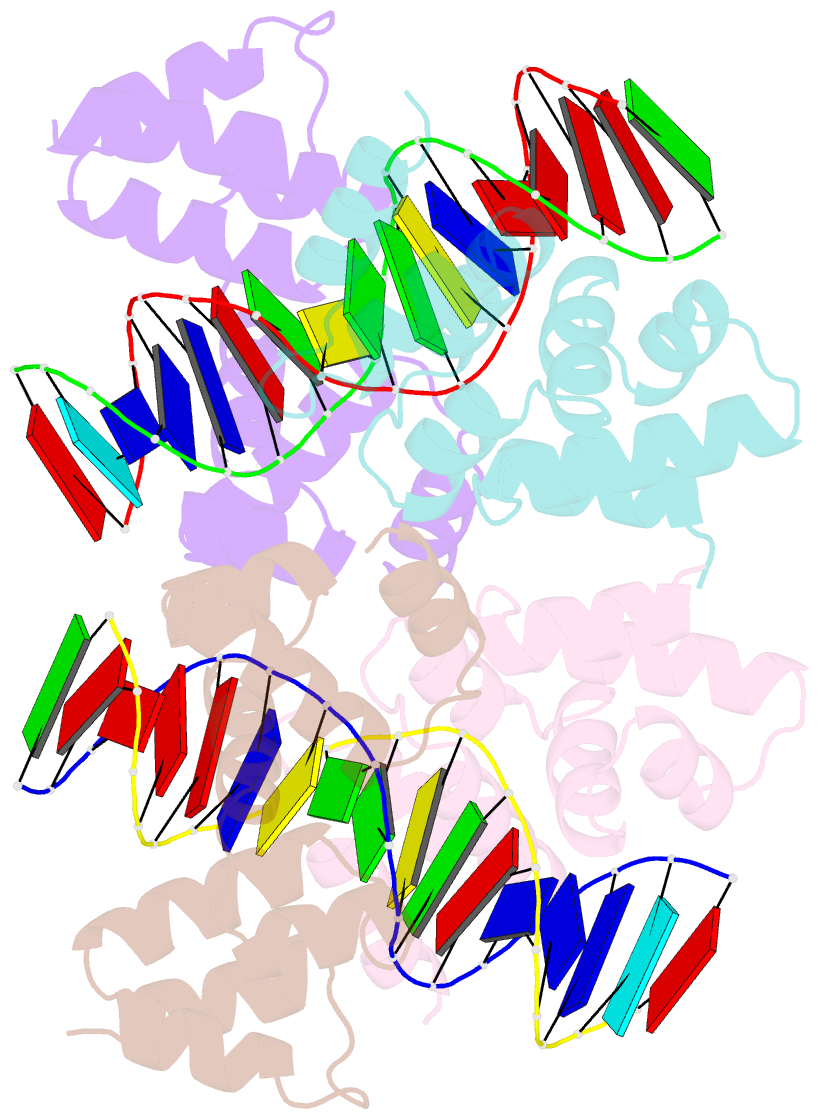
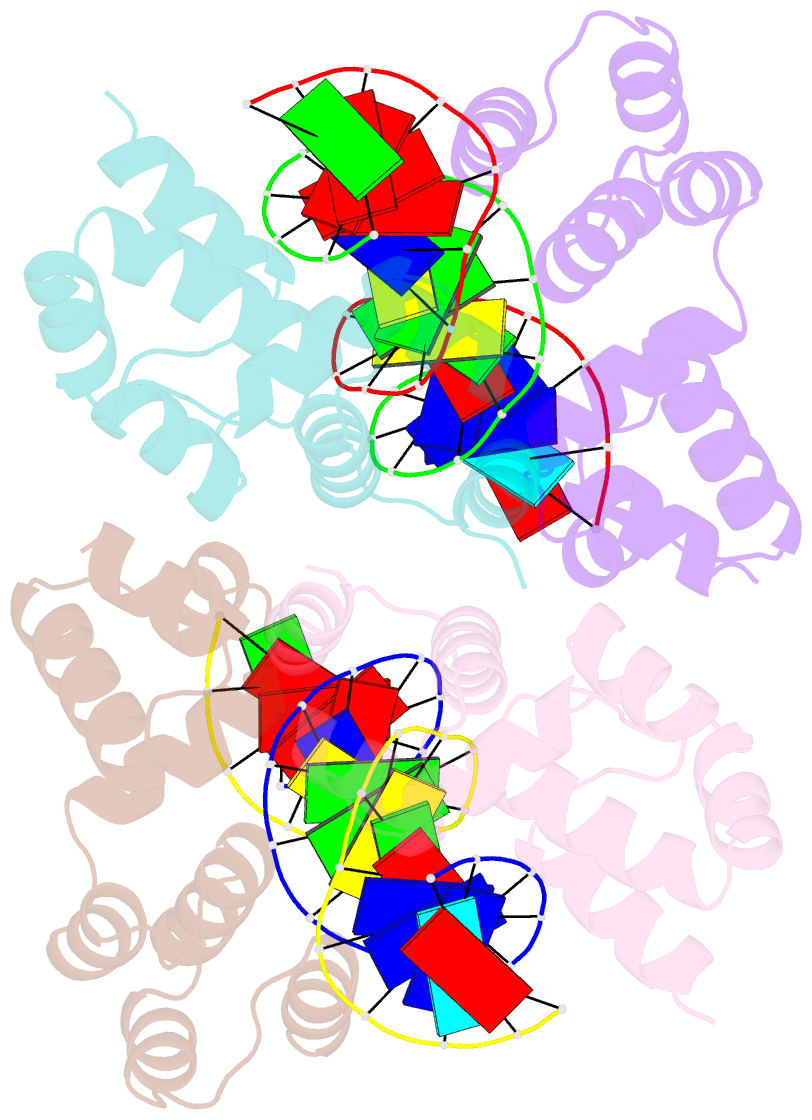
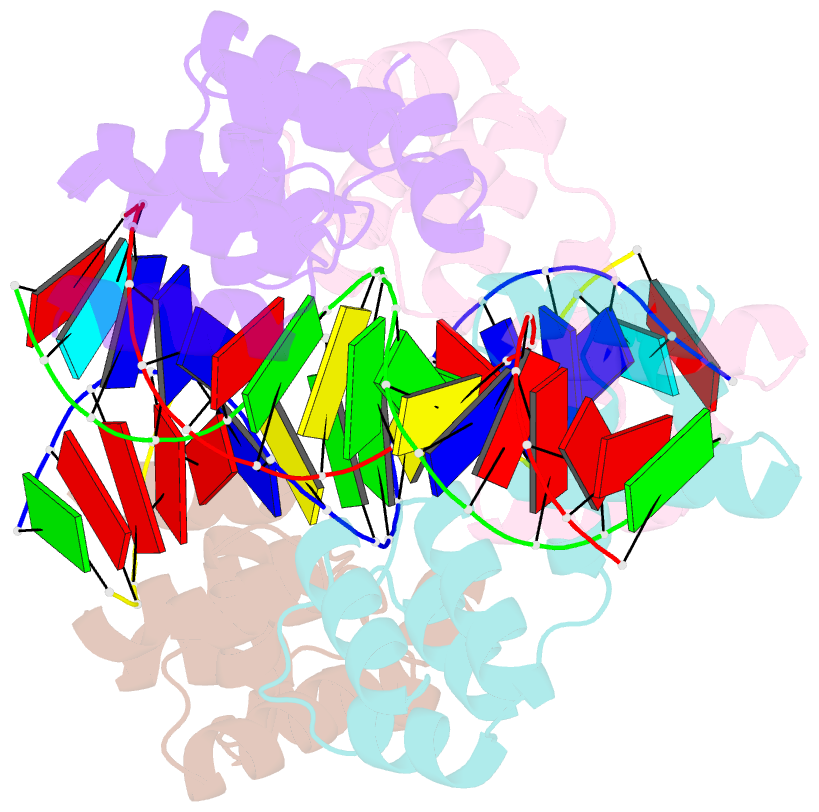
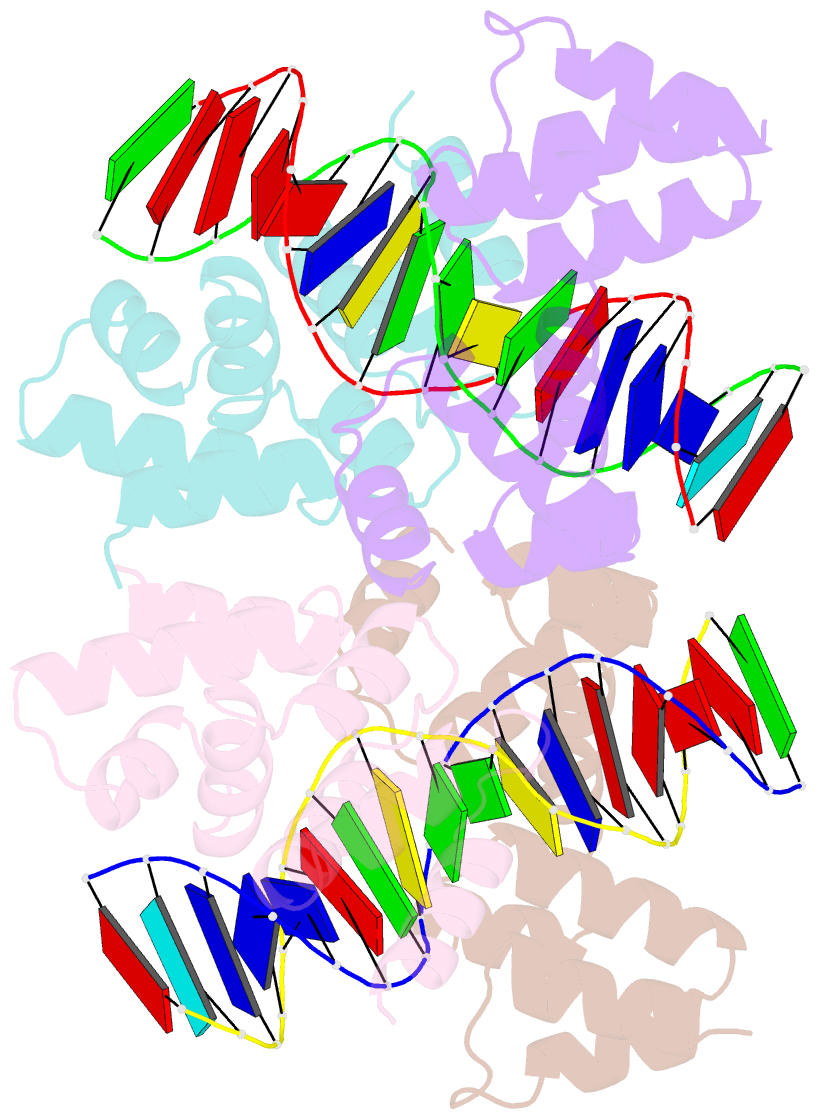
- Crystal structure of the DNA binding domain of korb in complex with the operator DNA
- Reference
- Khare D, Ziegelin G, Lanka E, Heinemann U (2004): "Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB." Nat.Struct.Mol.Biol., 11, 656-663. doi: 10.1038/nsmb773.
- Abstract
- The KorB protein of the broad-host-range plasmid RP4 acts as a multifunctional regulator of plasmid housekeeping genes, including those responsible for replication, maintenance and conjugation. Additionally, KorB functions as the ParB analog of the plasmid's partitioning system. The protein structure consists of eight helices, two of which belong to a predicted helix-turn-helix motif. Each half-site of the palindromic operator DNA binds one copy of the protein in the major groove. As confirmed by mutagenesis, recognition specificity is based mainly on two side chain interactions outside the helix-turn-helix motif with two bases next to the central base pair of the 13-base pair operator sequence. The surface of the KorB DNA-binding domain mirrors the overall acidity of KorB, whereas DNA binding occurs via a basic interaction surface. We present a model of KorB, including the structure of its dimerization domain, and discuss its interactions with the highly basic ParA homolog IncC.