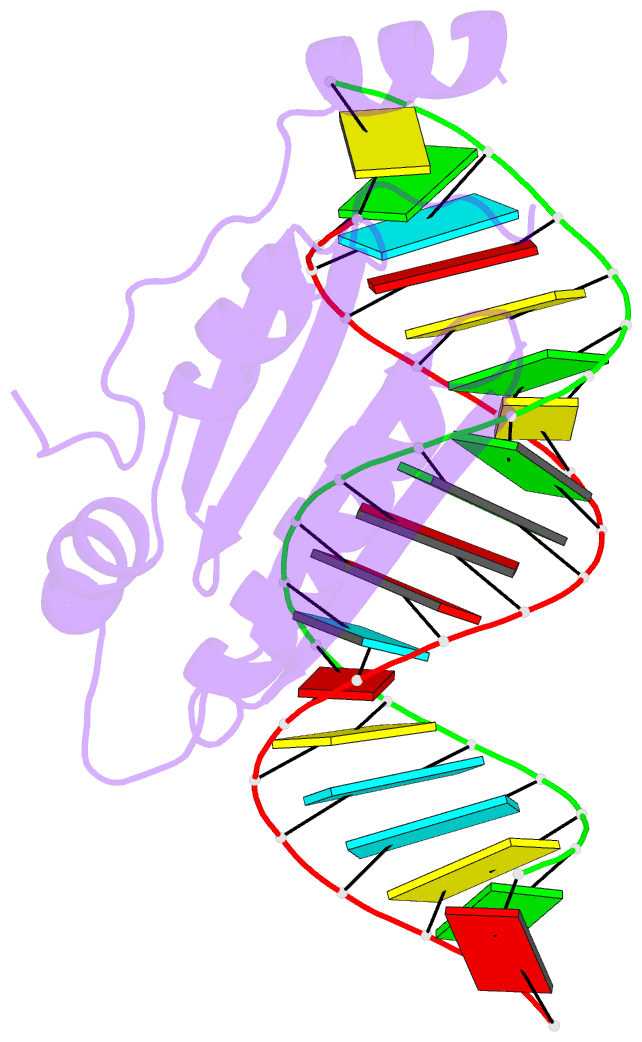
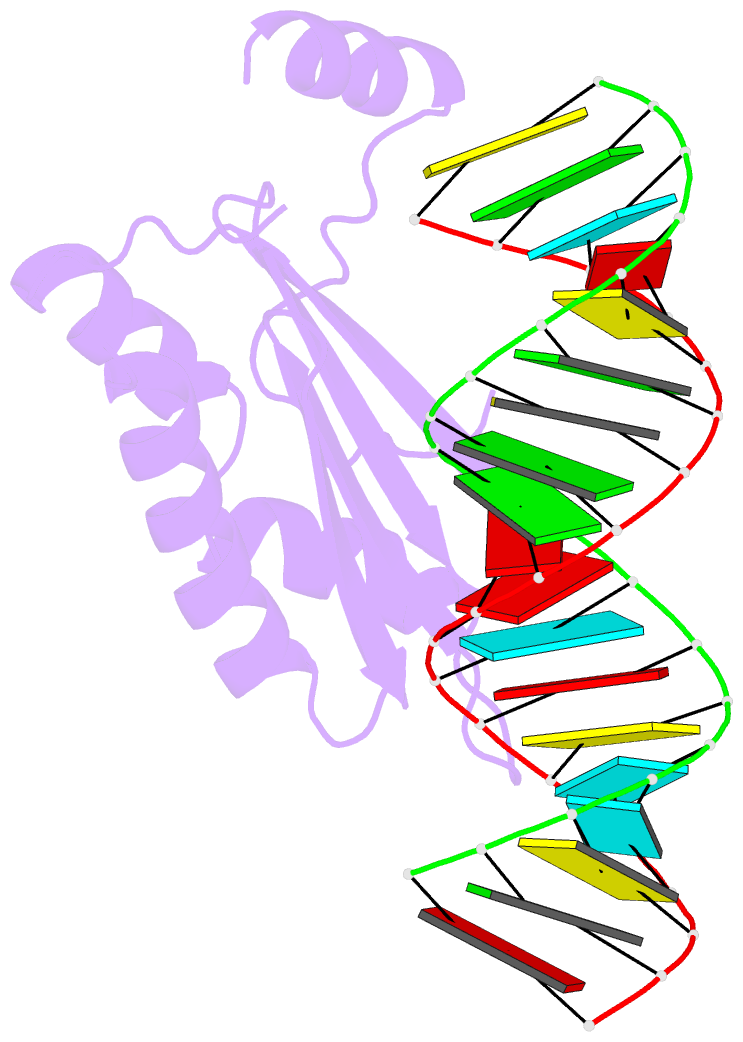
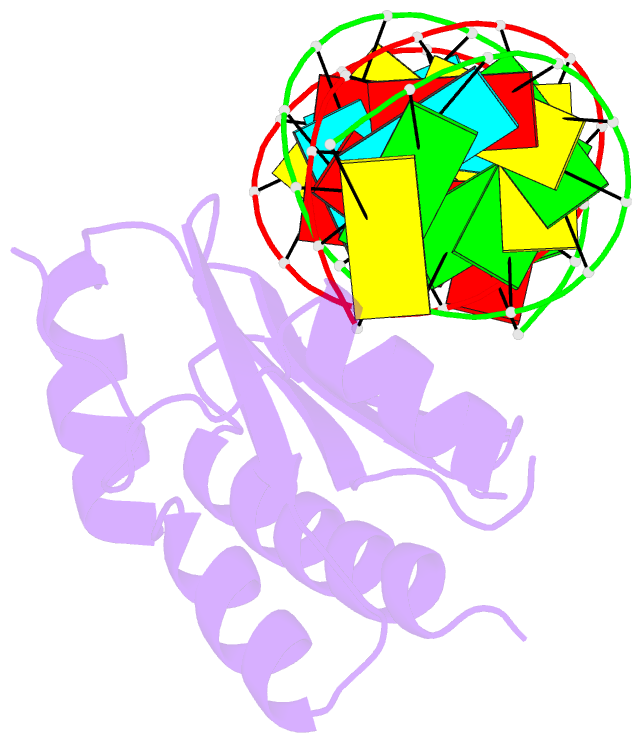
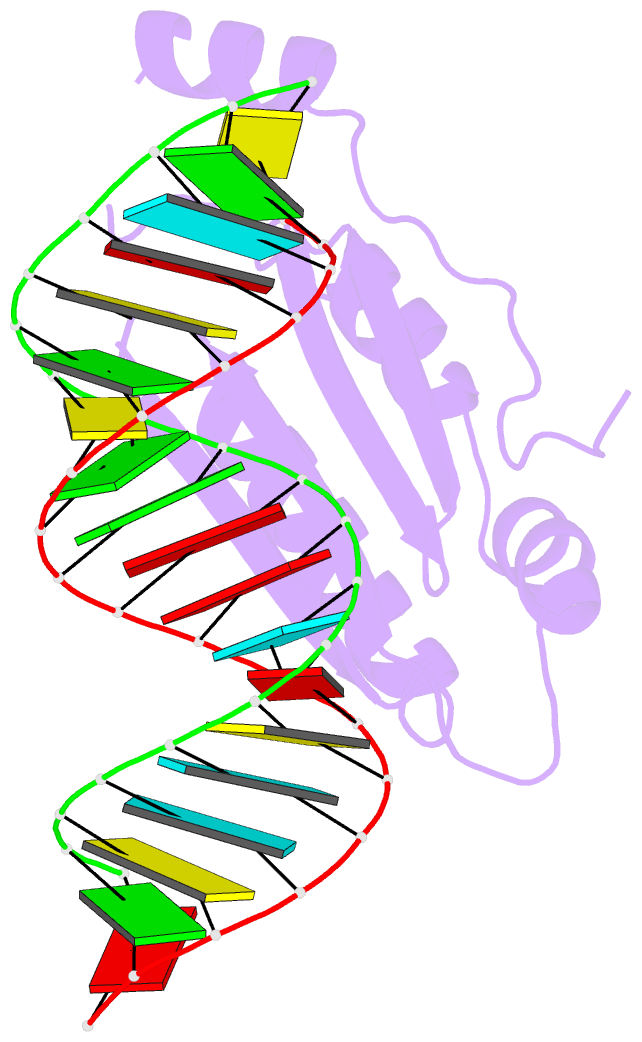
Summary information and primary citation
- PDB-id
- 1r9f; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- X-ray (1.85 Å)
- Summary
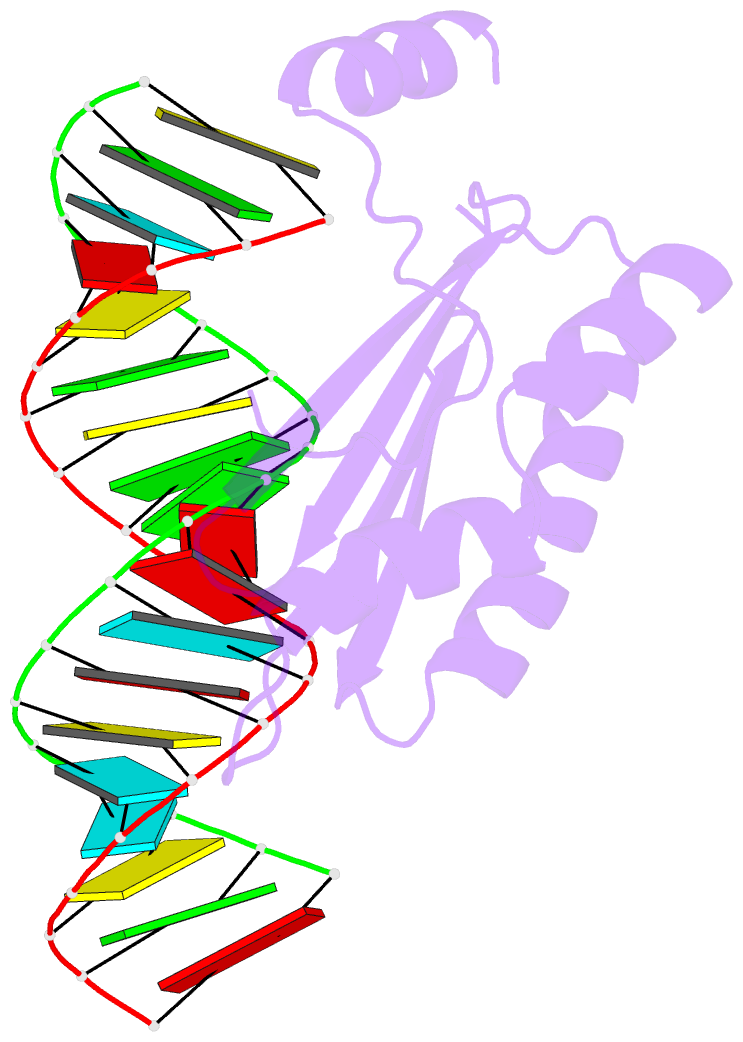
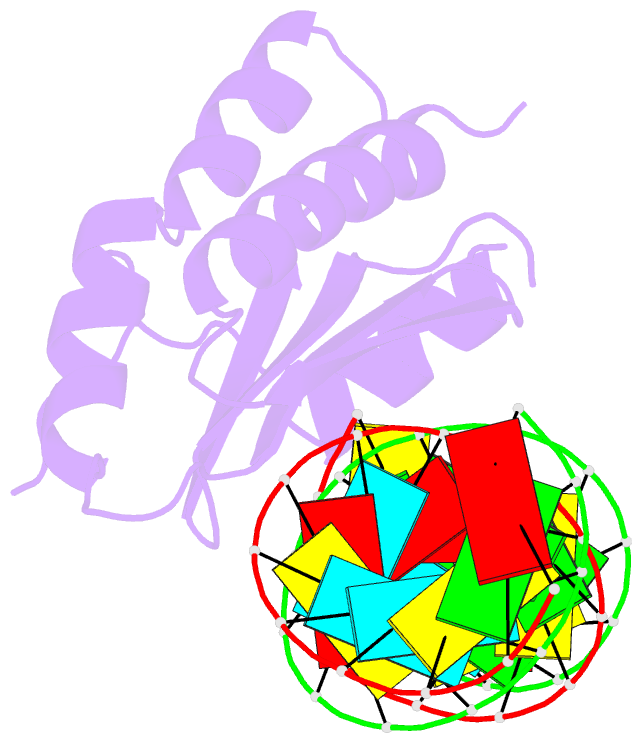
- Crystal structure of p19 complexed with 19-bp small interfering RNA
- Reference
- Ye K, Malinina L, Patel DJ (2003): "Recognition of small interfering RNA by a viral suppressor of RNA." Nature, 426, 874-878. doi: 10.1038/nature02213.
- Abstract
- RNA silencing (also known as RNA interference) is a conserved biological response to double-stranded RNA that regulates gene expression, and has evolved in plants as a defence against viruses. The response is mediated by small interfering RNAs (siRNAs), which guide the sequence-specific degradation of cognate messenger RNAs. As a counter-defence, many viruses encode proteins that specifically inhibit the silencing machinery. The p19 protein from the tombusvirus is such a viral suppressor of RNA silencing and has been shown to bind specifically to siRNA. Here, we report the 1.85-A crystal structure of p19 bound to a 21-nucleotide siRNA, where the 19-base-pair RNA duplex is cradled within the concave face of a continuous eight-stranded beta-sheet, formed across the p19 homodimer interface. Direct and water-mediated intermolecular contacts are restricted to the backbone phosphates and sugar 2'-OH groups, consistent with sequence-independent p19-siRNA recognition. Two alpha-helical 'reading heads' project from opposite ends of the p19 homodimer and position pairs of tryptophans for stacking over the terminal base pairs, thereby measuring and bracketing both ends of the siRNA duplex. Our structure provides an illustration of siRNA sequestering by a viral protein.