Summary information and primary citation
- PDB-id
- 1rh6; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (1.7 Å)
- Summary
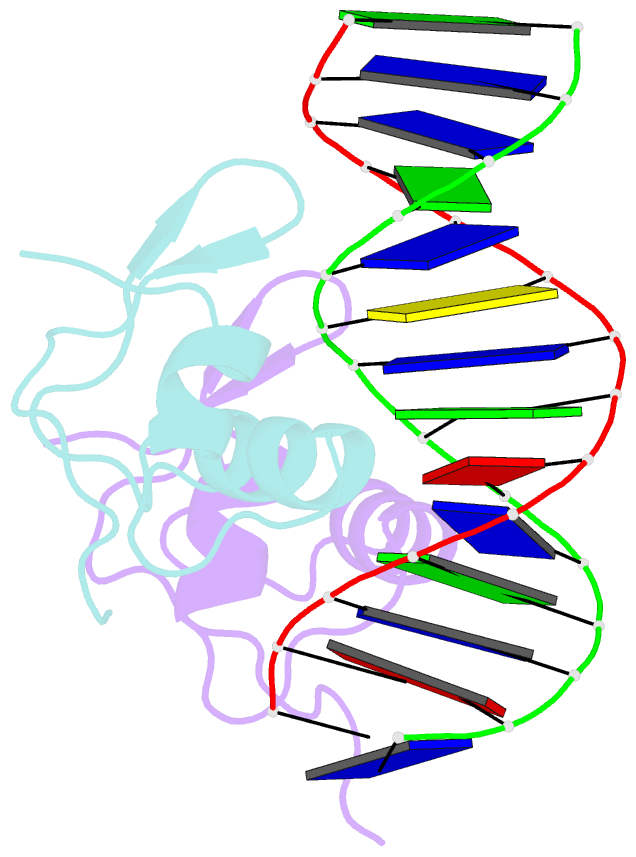
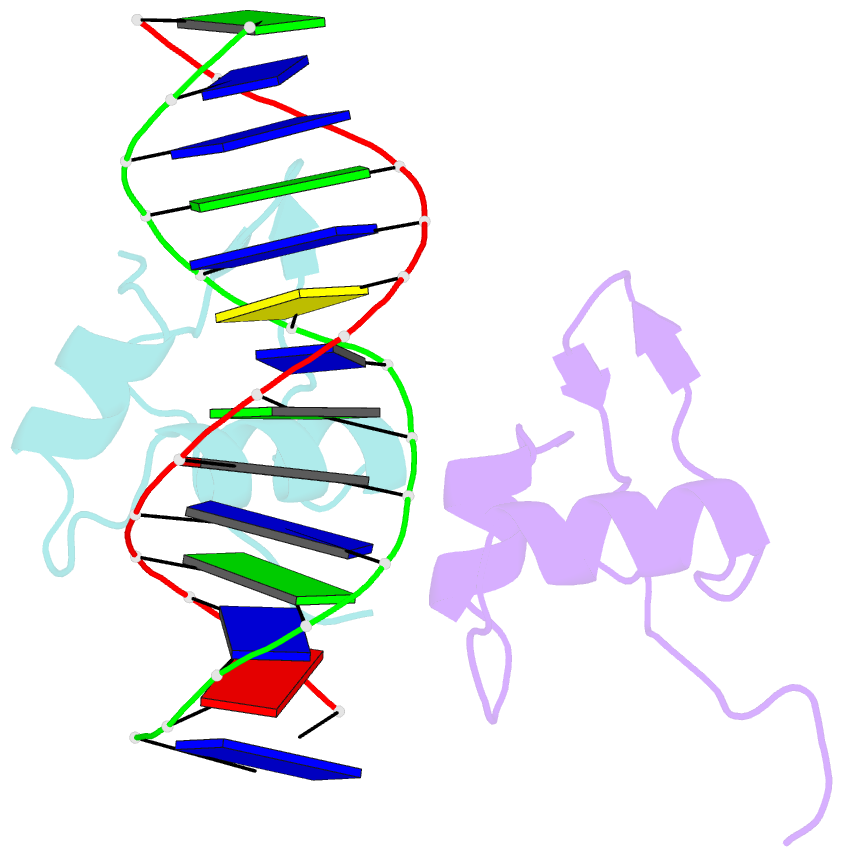
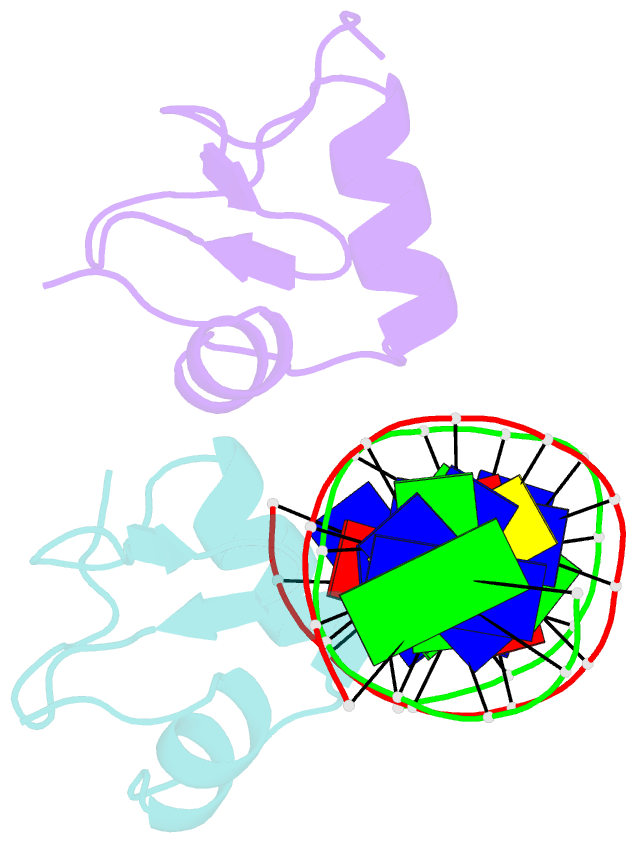
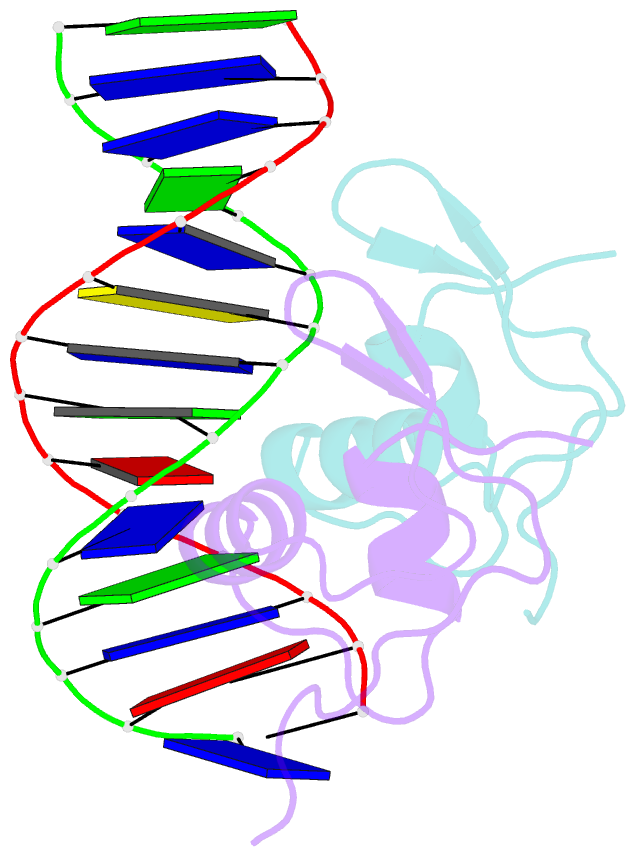
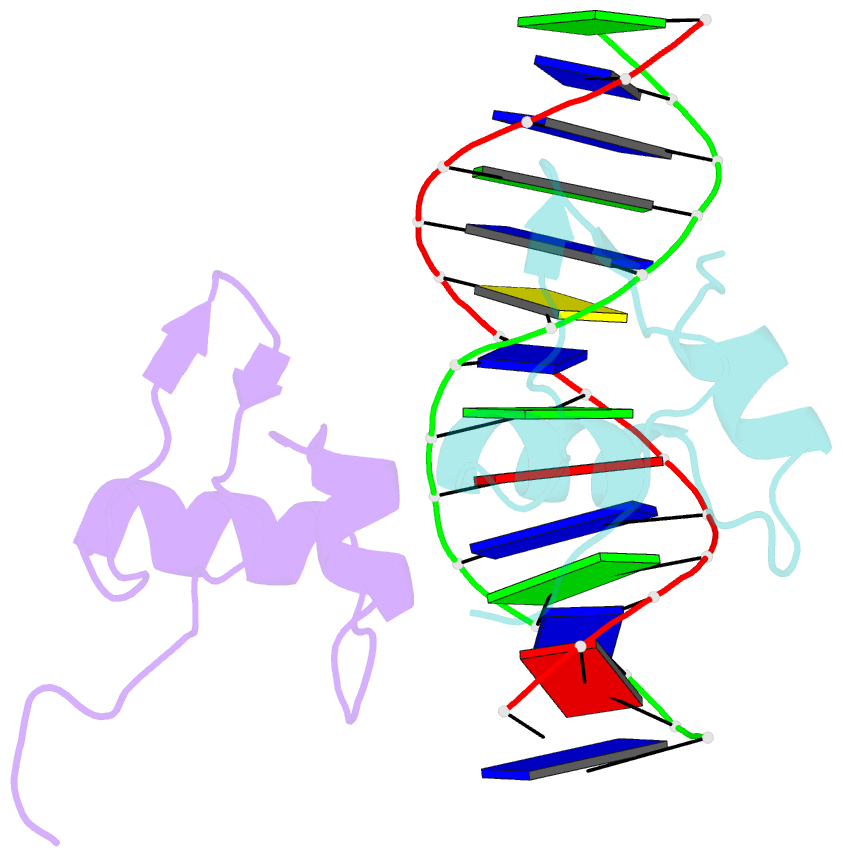
- Bacteriophage lambda excisionase (xis)-DNA complex
- Reference
- Sam MD, Cascio D, Johnson RC, Clubb RT (2004): "Crystal structure of the excisionase-DNA complex from bacteriophage lambda." J.Mol.Biol., 338, 229-240. doi: 10.1016/j.jmb.2004.02.053.
- Abstract
- The excisionase (Xis) protein from bacteriophage lambda is the best characterized member of a large family of recombination directionality factors that control integrase-mediated DNA rearrangements. It triggers phage excision by cooperatively binding to sites X1 and X2 within the phage, bending DNA significantly and recruiting the phage-encoded integrase (Int) protein to site P2. We have determined the co-crystal structure of Xis with its X2 DNA-binding site at 1.7A resolution. Xis forms a unique winged-helix motif that interacts with the major and minor grooves of its binding site using an alpha-helix and an ordered beta-hairpin (wing), respectively. Recognition is achieved through an elaborate water-mediated hydrogen-bonding network at the major groove interface, while the preformed hairpin forms largely non-specific interactions with the minor groove. The structure of the complex provides insights into how Xis recruits Int cooperatively, and suggests a plausible mechanism by which it may distort longer DNA fragments significantly. It reveals a surface on the protein that is likely to mediate Xis-Xis interactions required for its cooperative binding to DNA.