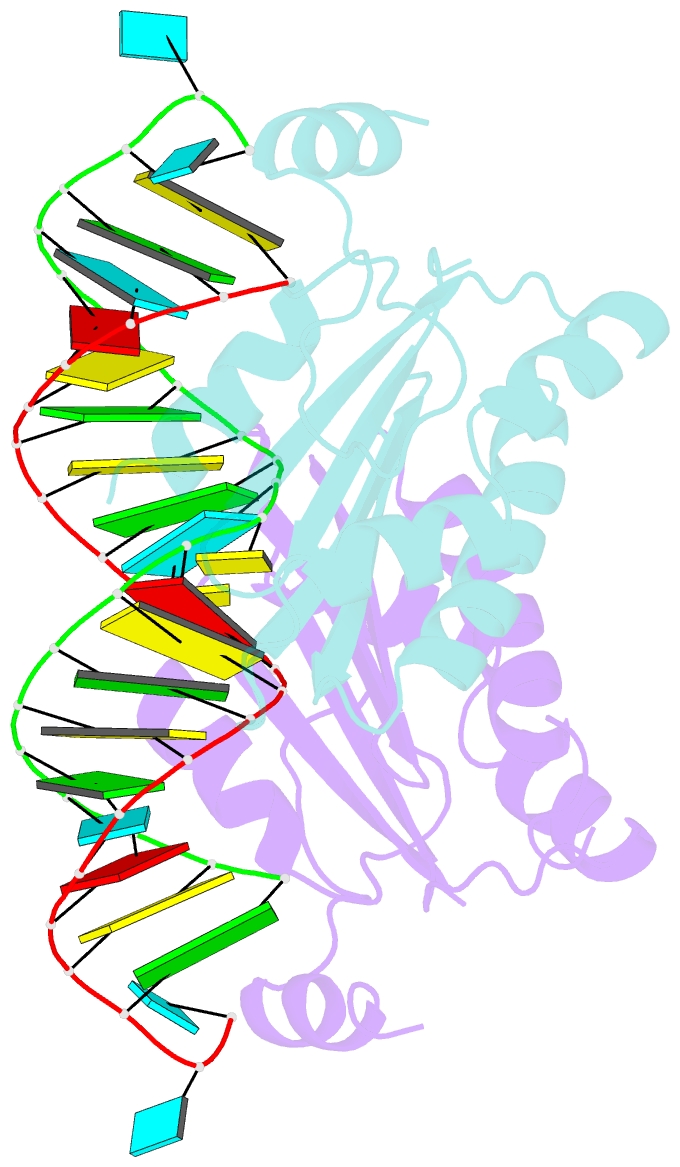
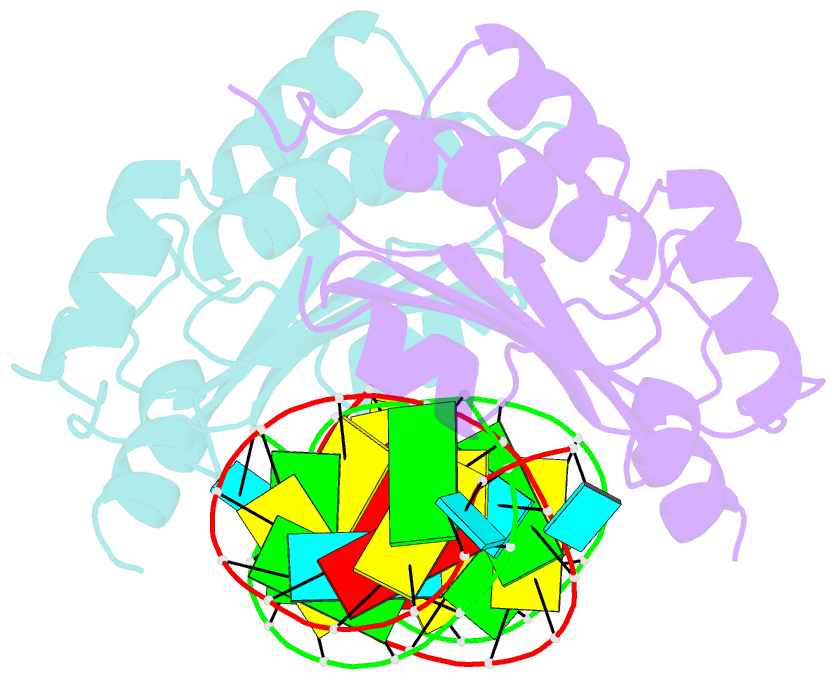
Summary information and primary citation
- PDB-id
- 1rpu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (2.5 Å)
- Summary
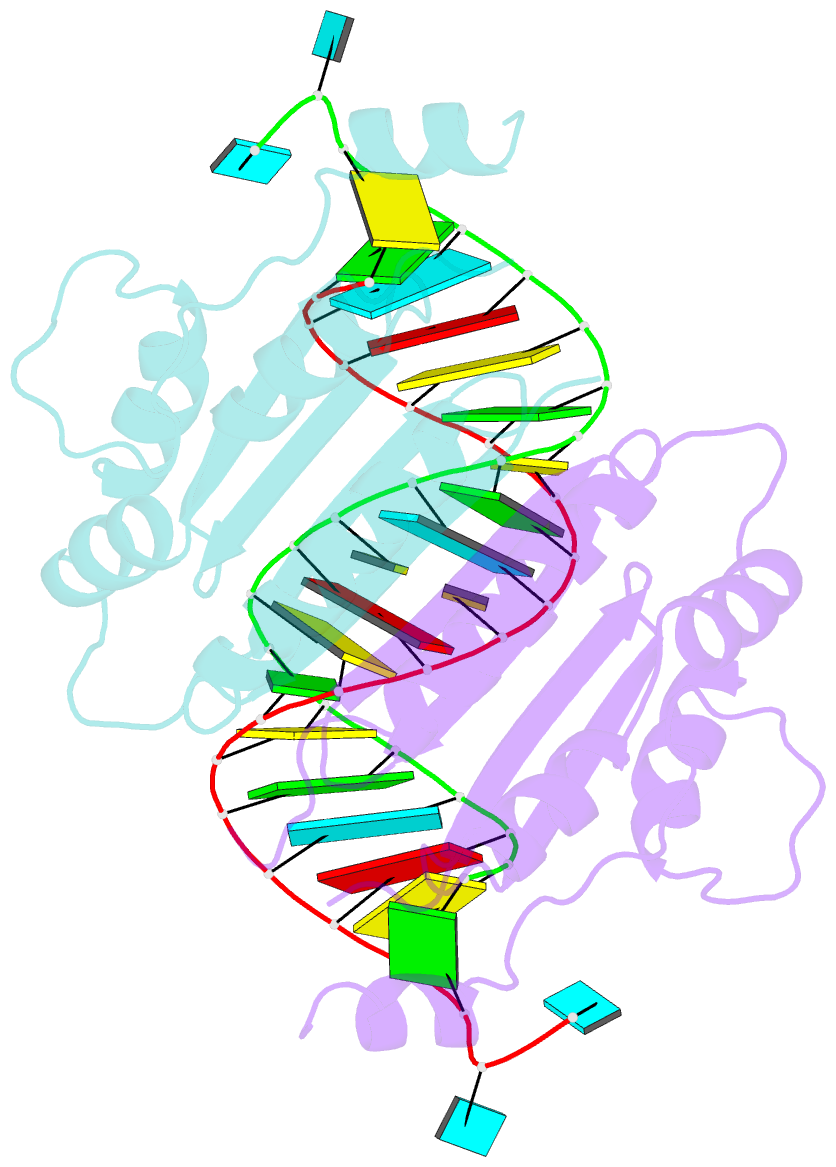
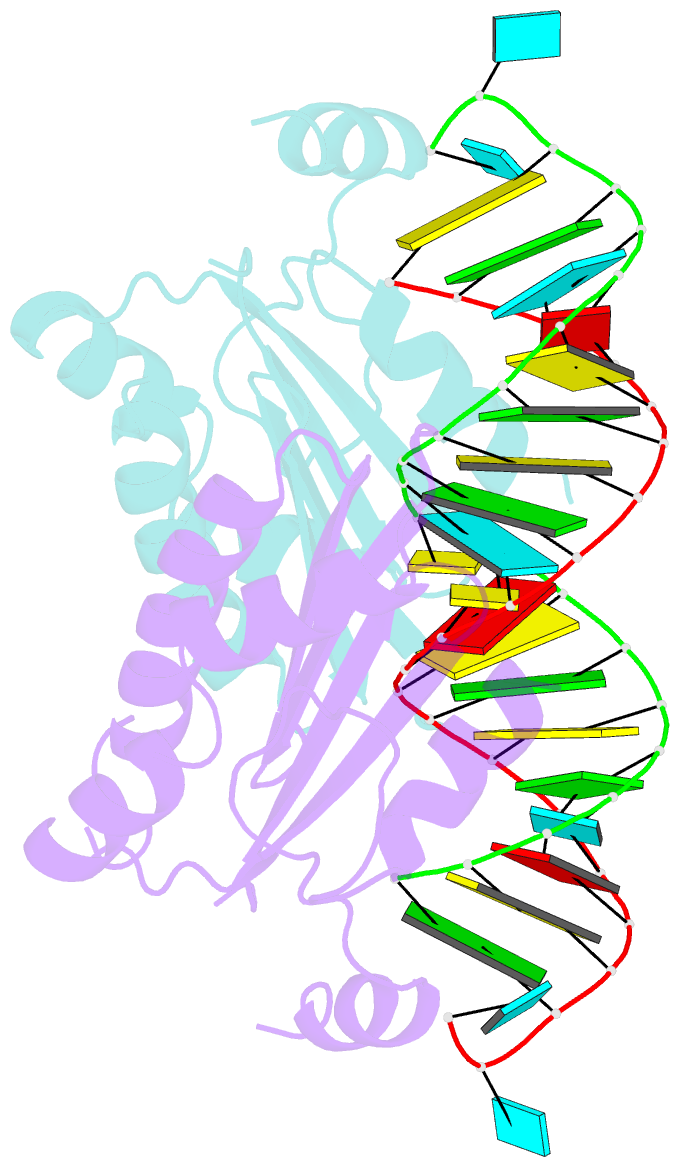
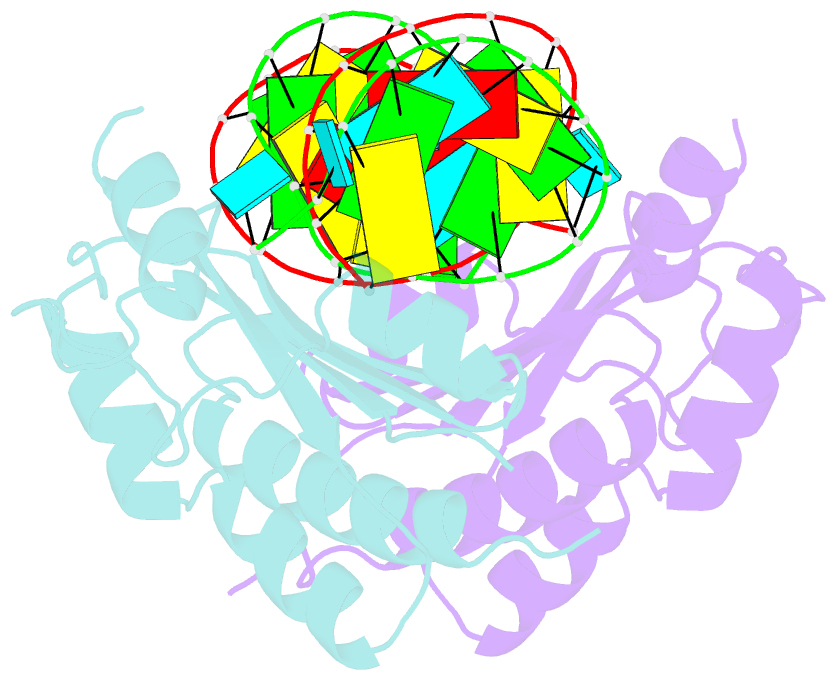
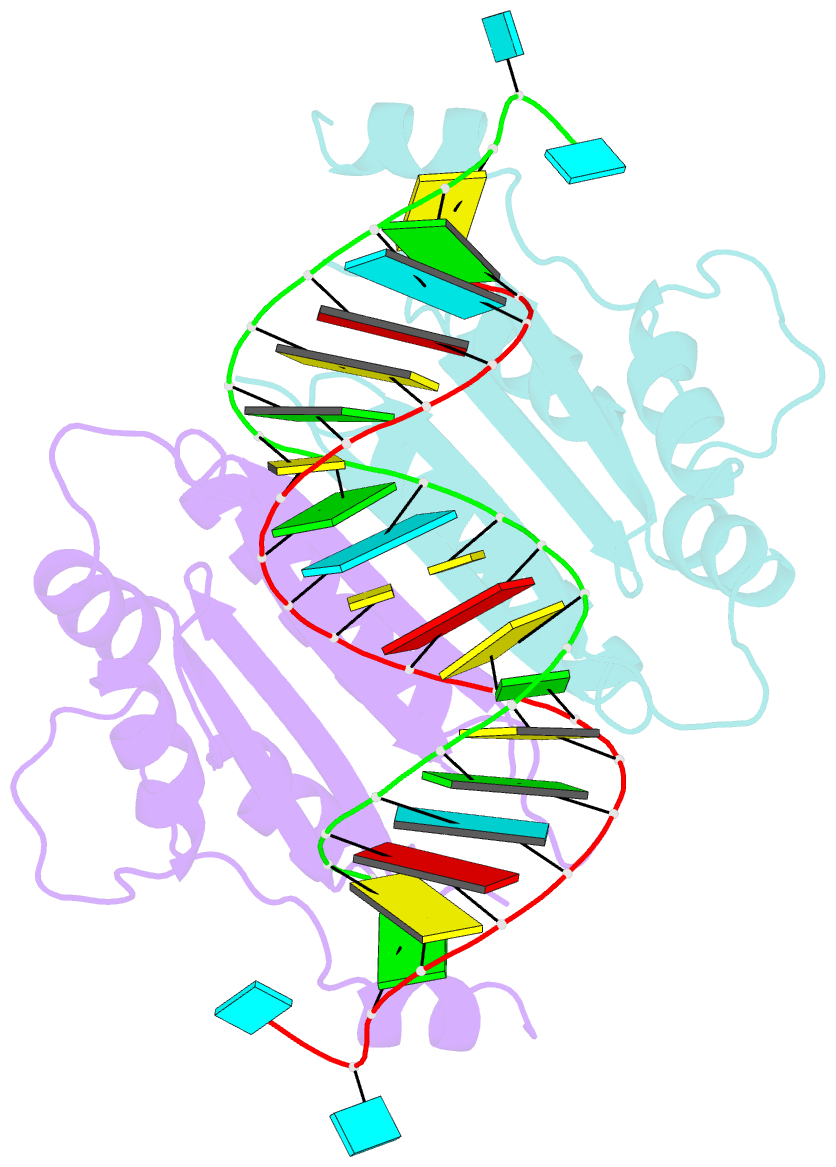
- Crystal structure of cirv p19 bound to sirna
- Reference
- Vargason JM, Szittya G, Burgyan J, Hall TMT (2003): "Size selective recognition of siRNA by an RNA silencing suppressor." Cell(Cambridge,Mass.), 115, 799-811. doi: 10.1016/S0092-8674(03)00984-X.
- Abstract
- RNA silencing in plants likely exists as a defense mechanism against molecular parasites such as RNA viruses, retrotransposons, and transgenes. As a result, many plant viruses have adapted mechanisms to evade and suppress gene silencing. Tombusviruses express a 19 kDa protein (p19), which has been shown to suppress RNA silencing in vivo and bind silencing-generated and synthetic small interfering RNAs (siRNAs) in vitro. Here we report the 2.5 A crystal structure of p19 from the Carnation Italian ringspot virus (CIRV) bound to a 21 nt siRNA and demonstrate in biochemical and in vivo assays that CIRV p19 protein acts as a molecular caliper to specifically select siRNAs based on the length of the duplex region of the RNA.