Summary information and primary citation
- PDB-id
- 1rta; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.5 Å)
- Summary
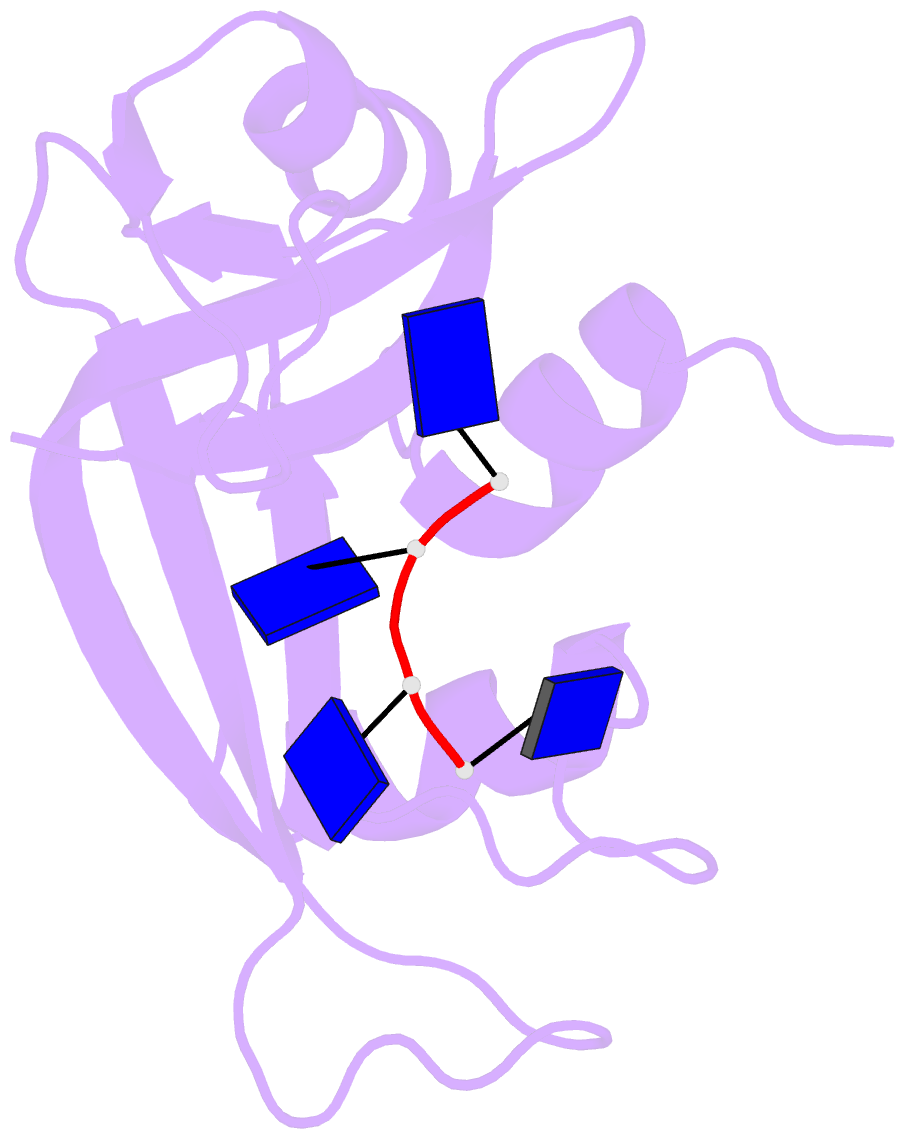
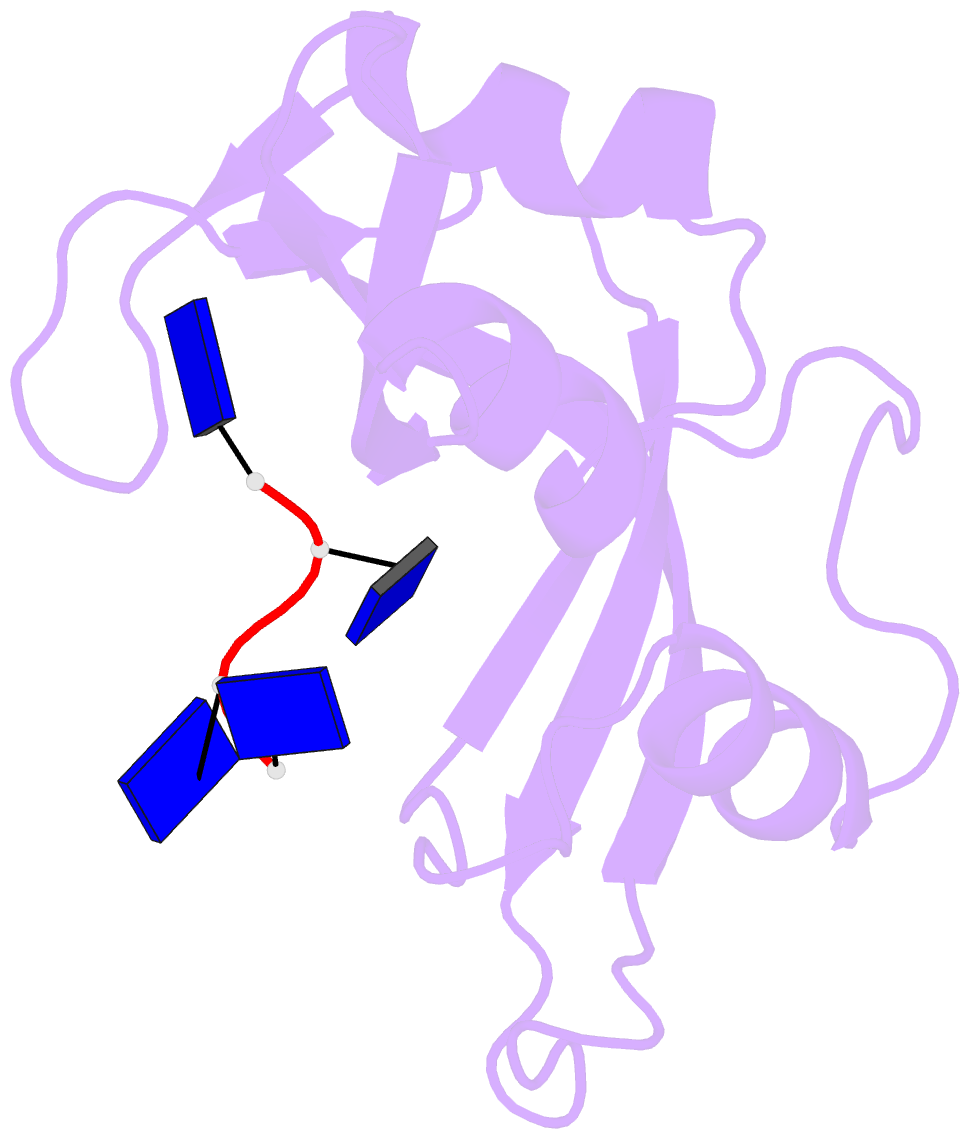
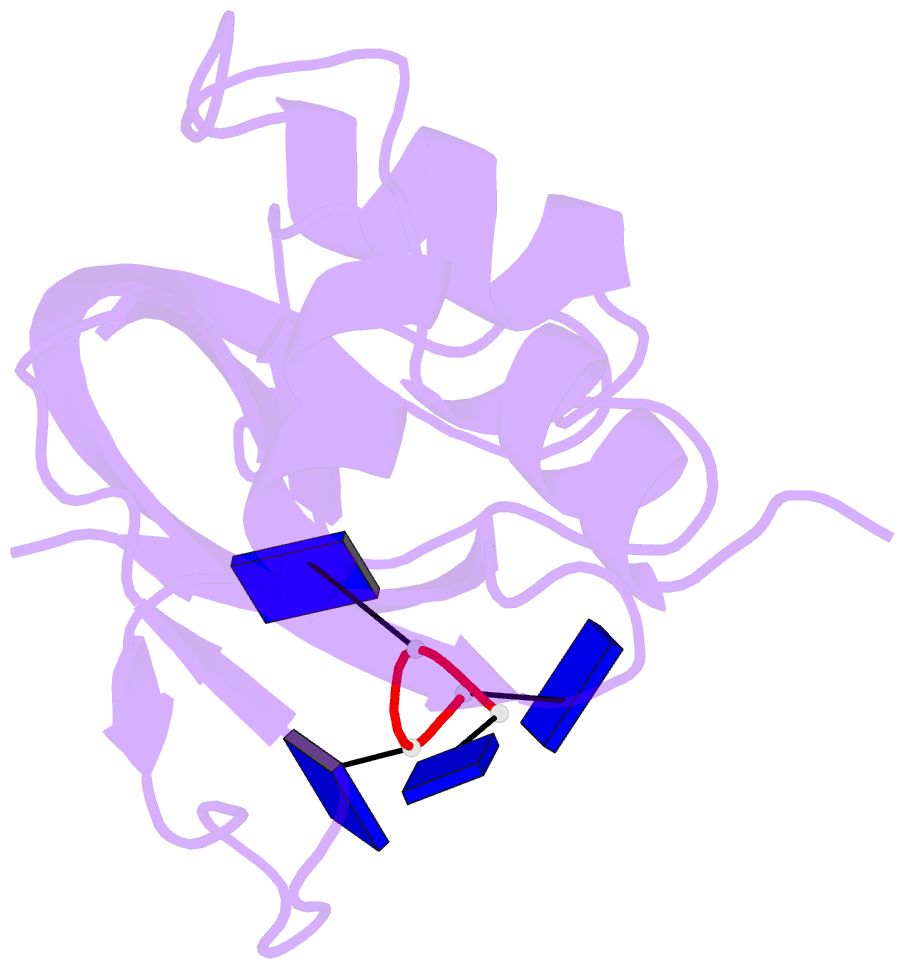
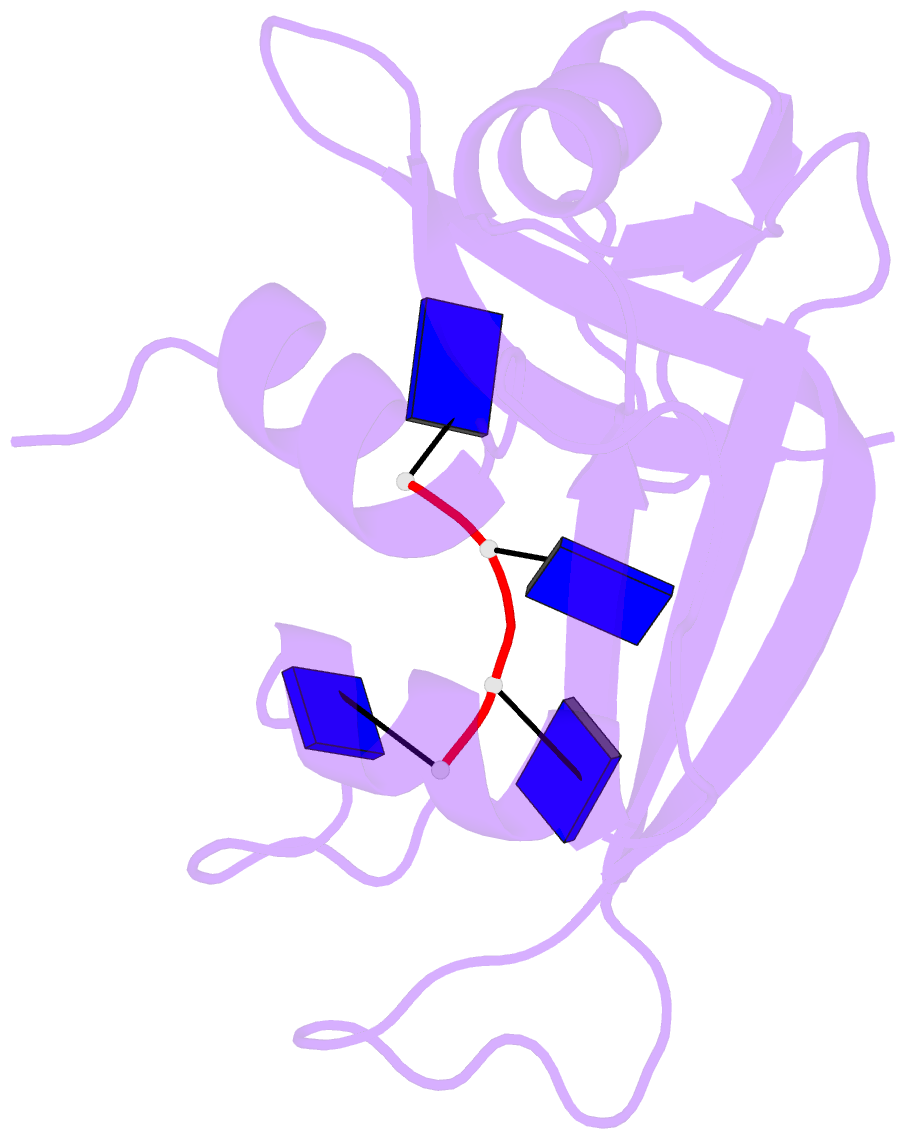
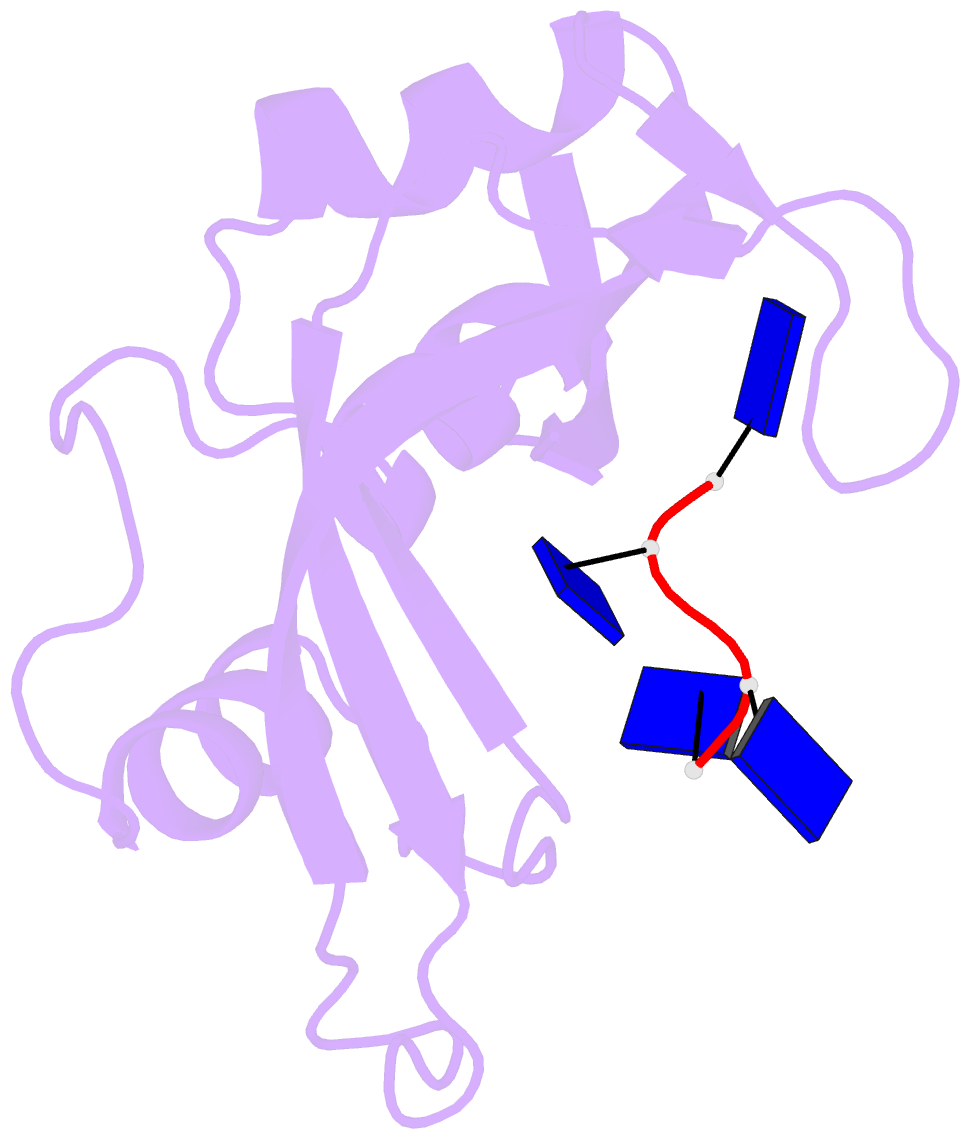
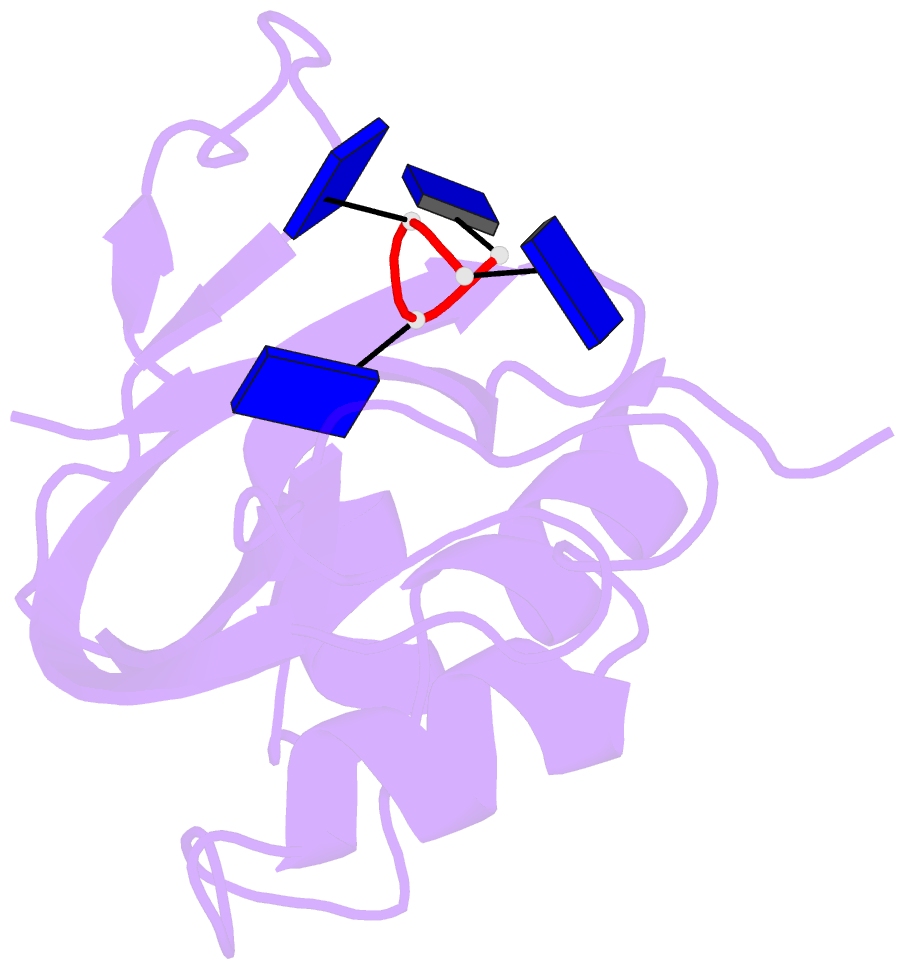
- Crystal structure disposition of thymidylic acid tetramer in complex with ribonuclease a
- Reference
- Birdsall DL, McPherson A (1992): "Crystal structure disposition of thymidylic acid tetramer in complex with ribonuclease A." J.Biol.Chem., 267, 22230-22236.
- Abstract
- The crystal structure of ribonuclease A with bound thymidylic acid tetramer is reported at 2.5-A resolution. The diffusion of the tetramer into native orthorhombic crystals of the ribonuclease allows for the formation of a structurally stable complex where the single-stranded nucleic acid enters and leaves the enzyme's catalytic region in a persistent 5'-3' direction. The binding of the tetramer to the enzyme's surface is facilitated and mediated by electrostatic interactions between basic protein residues and nucleotide phosphates. Two pyrimidine nucleotides are bound to the enzyme's active site in a manner similar to that observed for other complexes between ribonuclease A and nucleic acid oligomers.