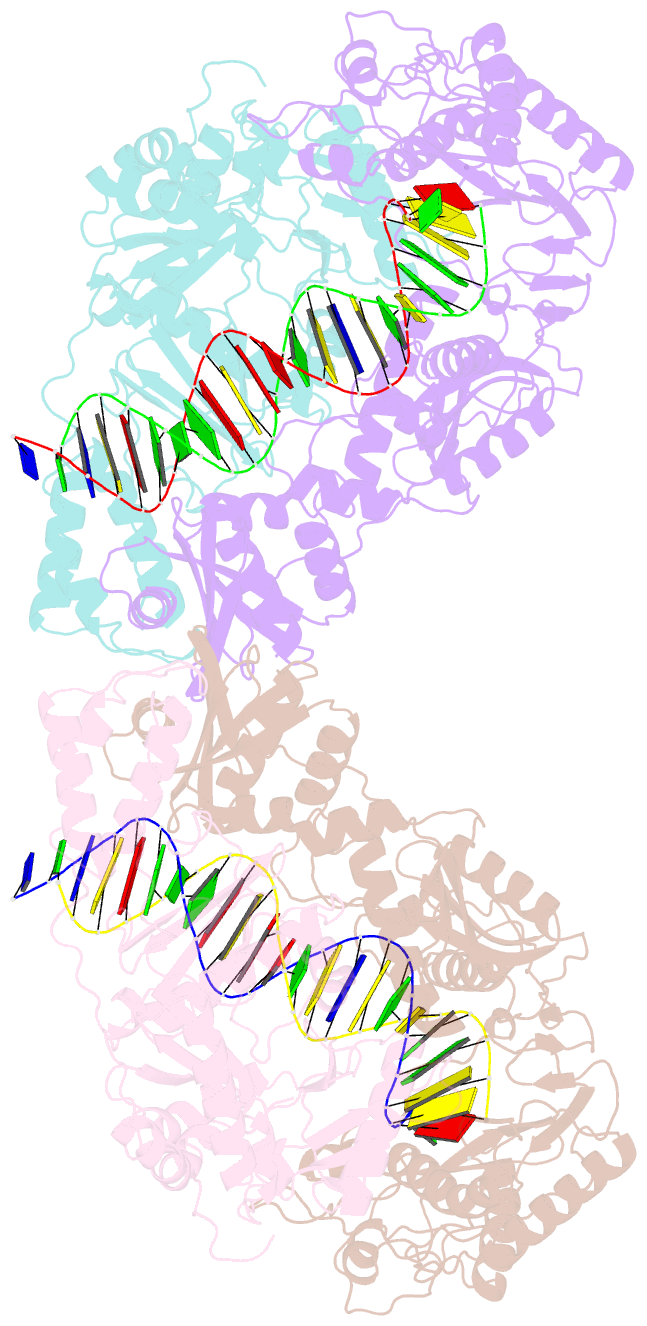
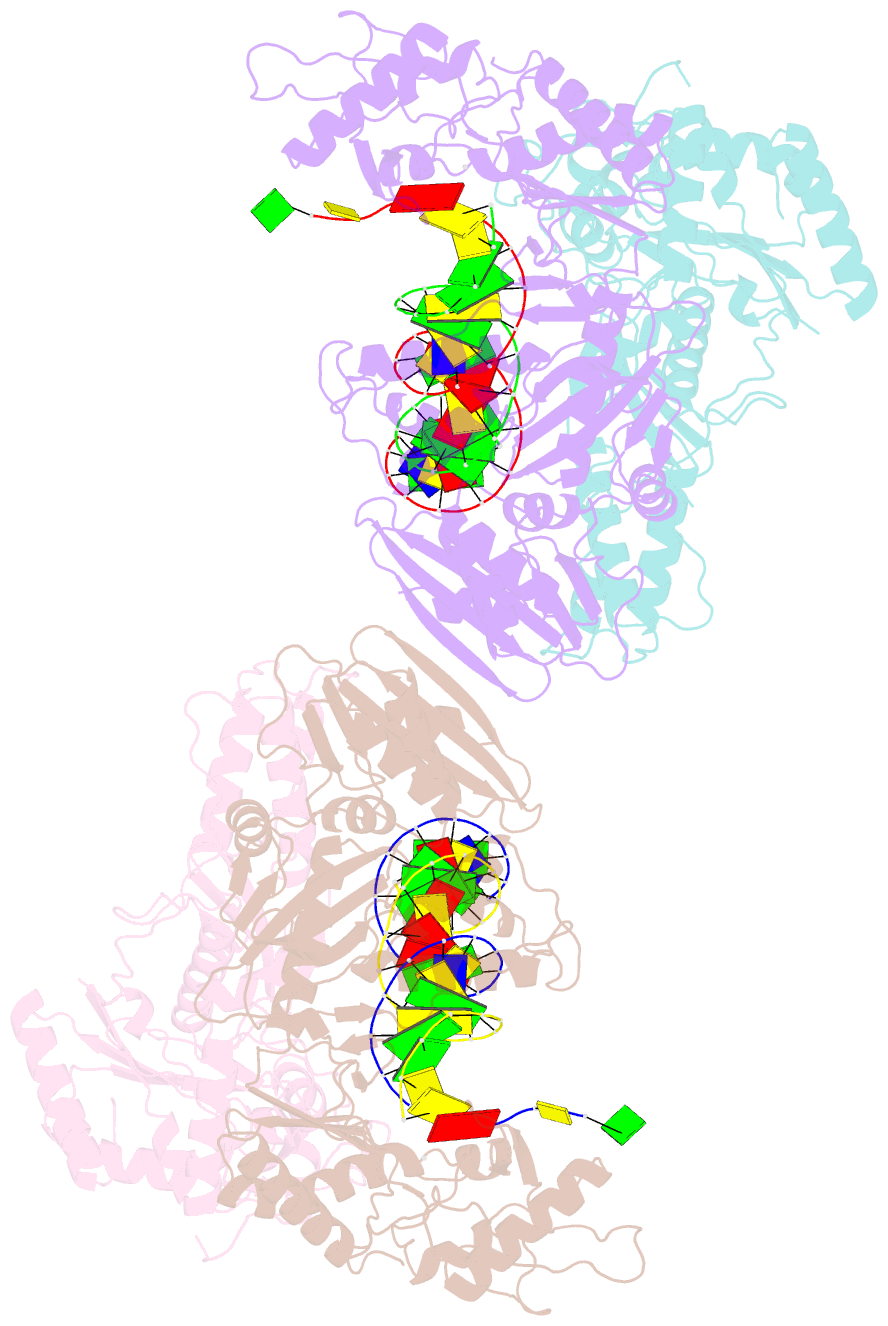
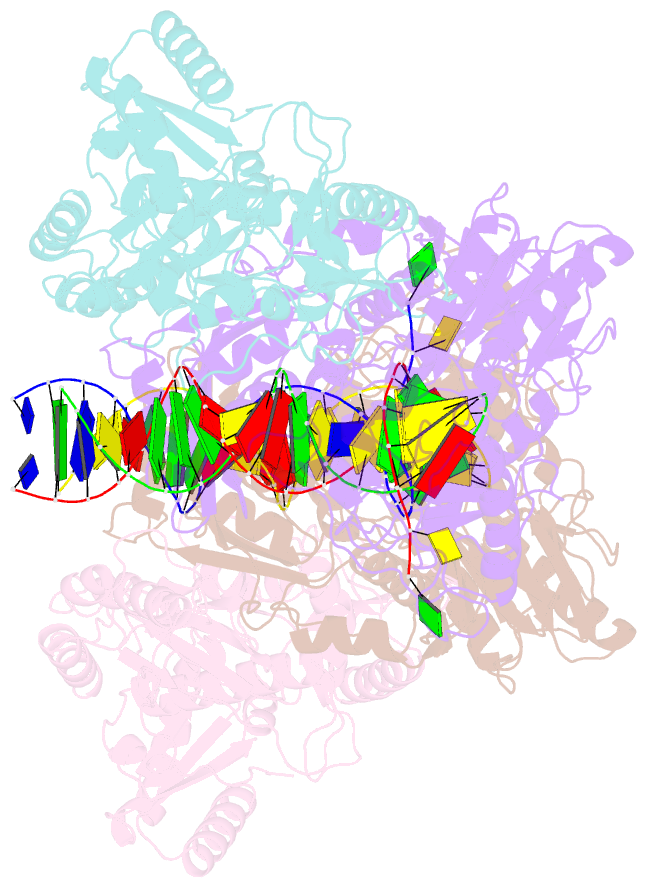
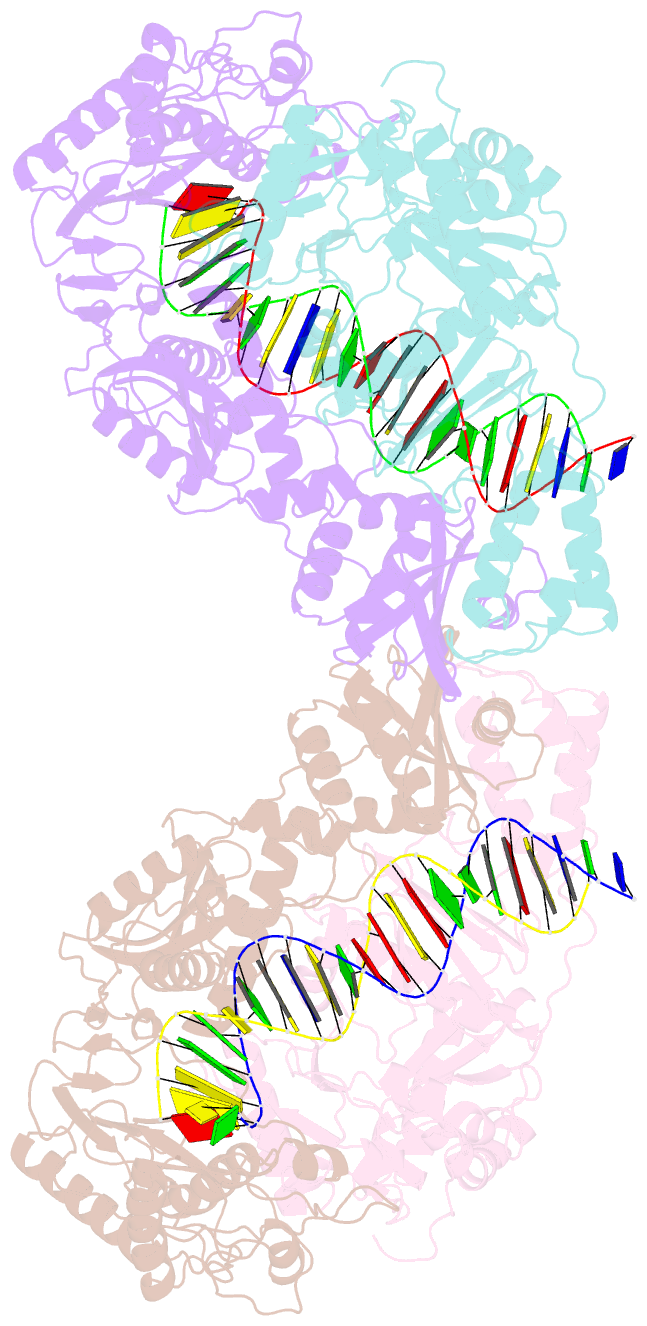
Summary information and primary citation
- PDB-id
- 1rtd; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (3.2 Å)
- Summary
- Structure of a catalytic complex of hiv-1 reverse transcriptase: implications for nucleoside analog drug resistance
- Reference
- Huang H, Chopra R, Verdine GL, Harrison SC (1998): "Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance." Science, 282, 1669-1675. doi: 10.1126/science.282.5394.1669.
- Abstract
- A combinatorial disulfide cross-linking strategy was used to prepare a stalled complex of human immunodeficiency virus-type 1 (HIV-1) reverse transcriptase with a DNA template:primer and a deoxynucleoside triphosphate (dNTP), and the crystal structure of the complex was determined at a resolution of 3.2 angstroms. The presence of a dideoxynucleotide at the 3'-primer terminus allows capture of a state in which the substrates are poised for attack on the dNTP. Conformational changes that accompany formation of the catalytic complex produce distinct clusters of the residues that are altered in viruses resistant to nucleoside analog drugs. The positioning of these residues in the neighborhood of the dNTP helps to resolve some long-standing puzzles about the molecular basis of resistance. The resistance mutations are likely to influence binding or reactivity of the inhibitors, relative to normal dNTPs, and the clustering of the mutations correlates with the chemical structure of the drug.