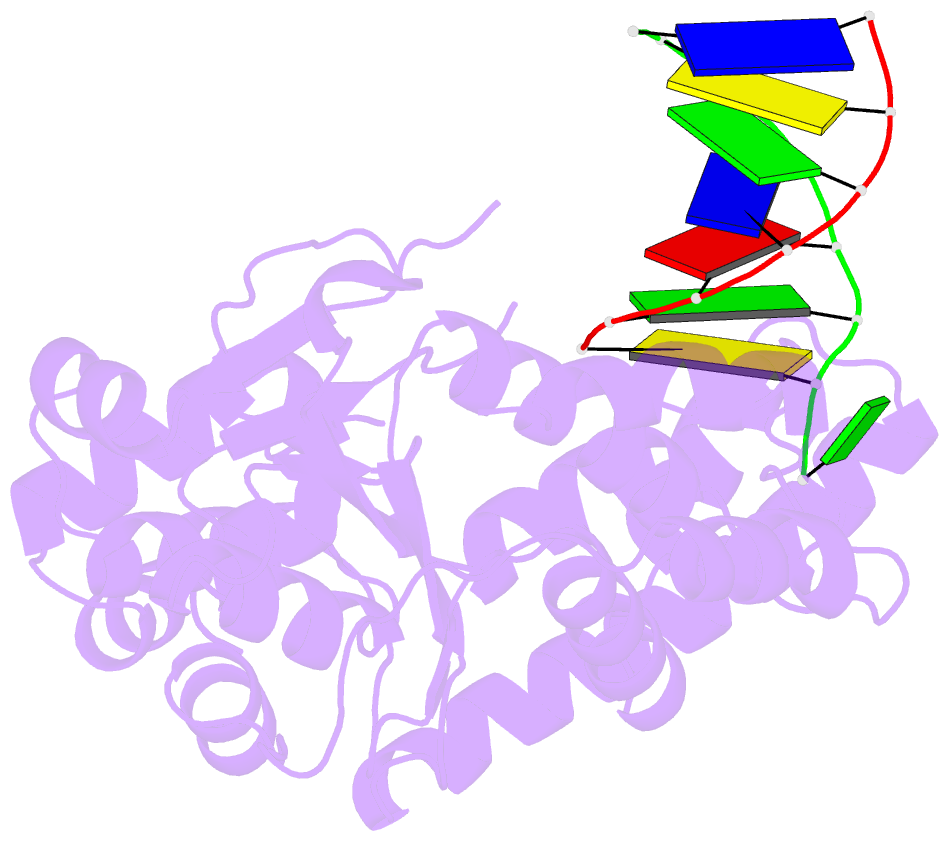
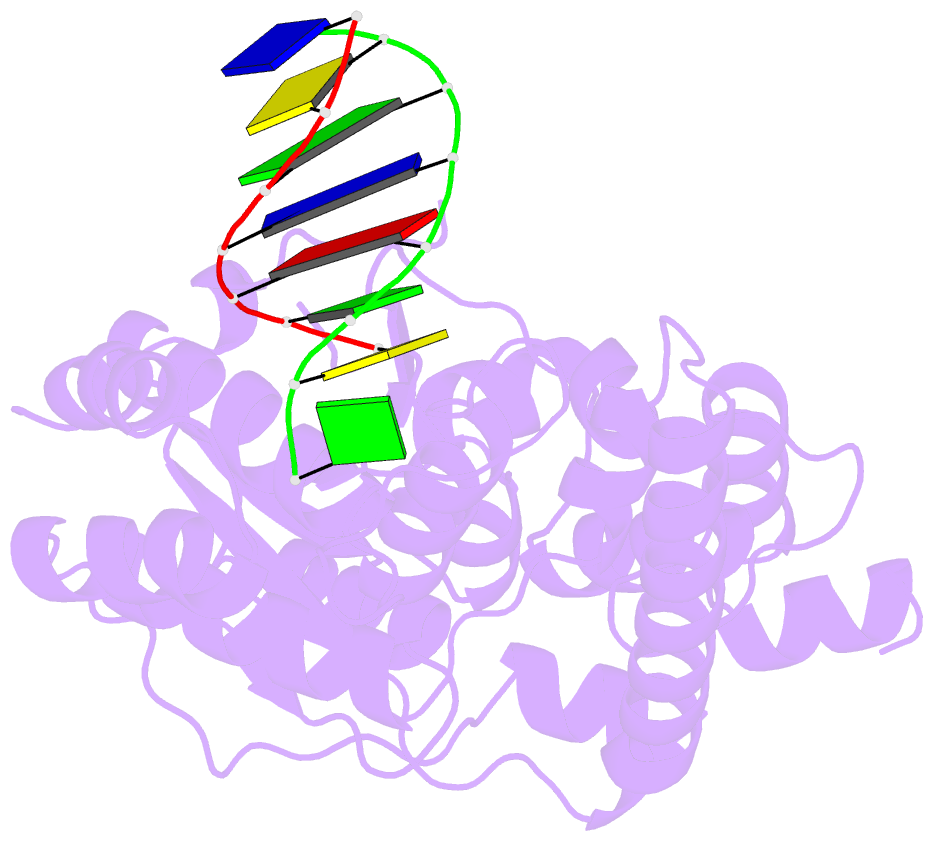
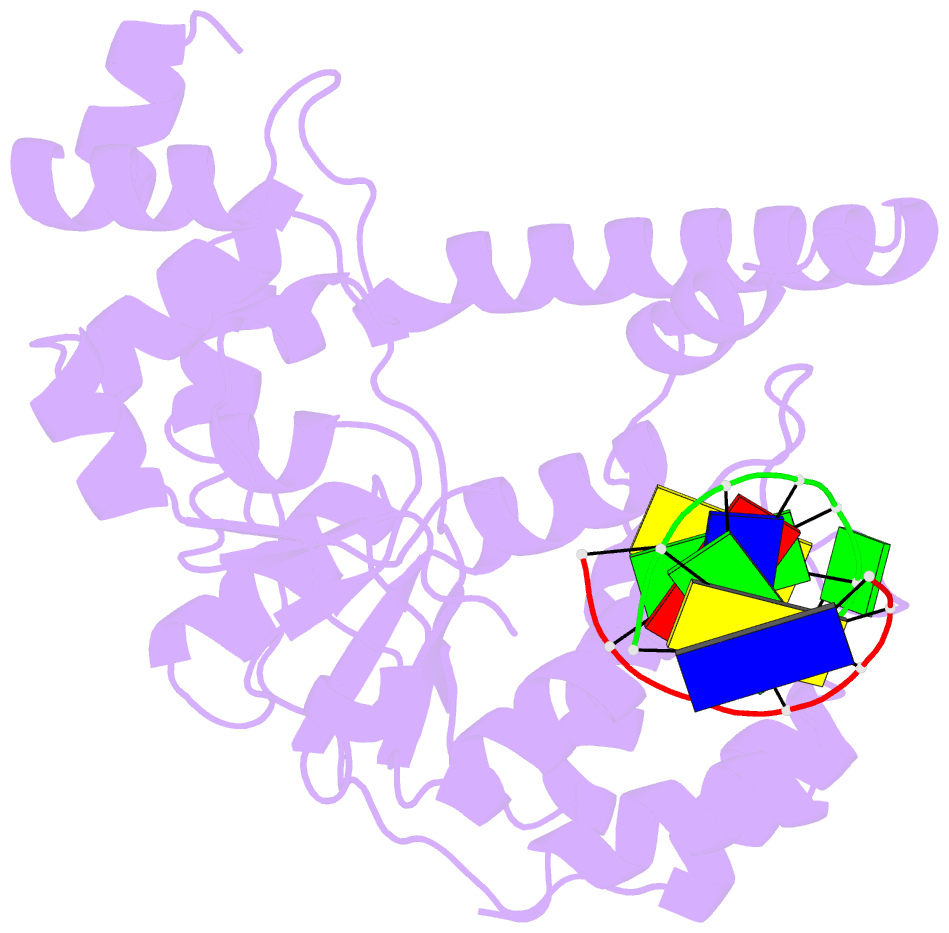
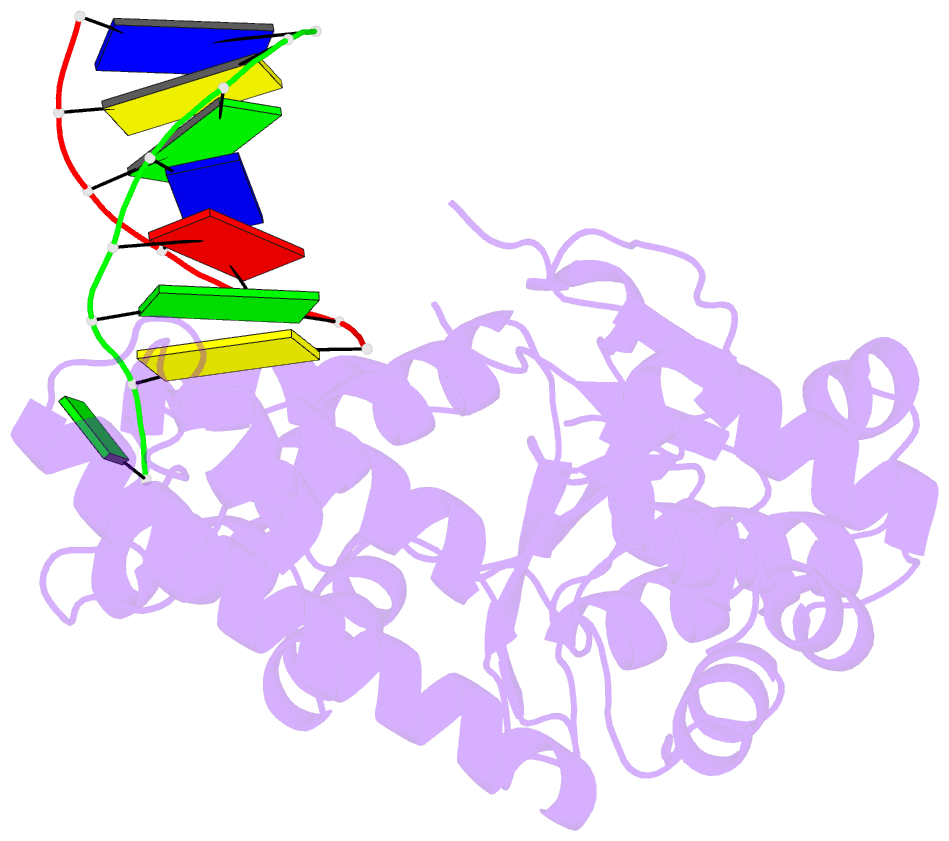
Summary information and primary citation
- PDB-id
- 1rxw; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.0 Å)
- Summary
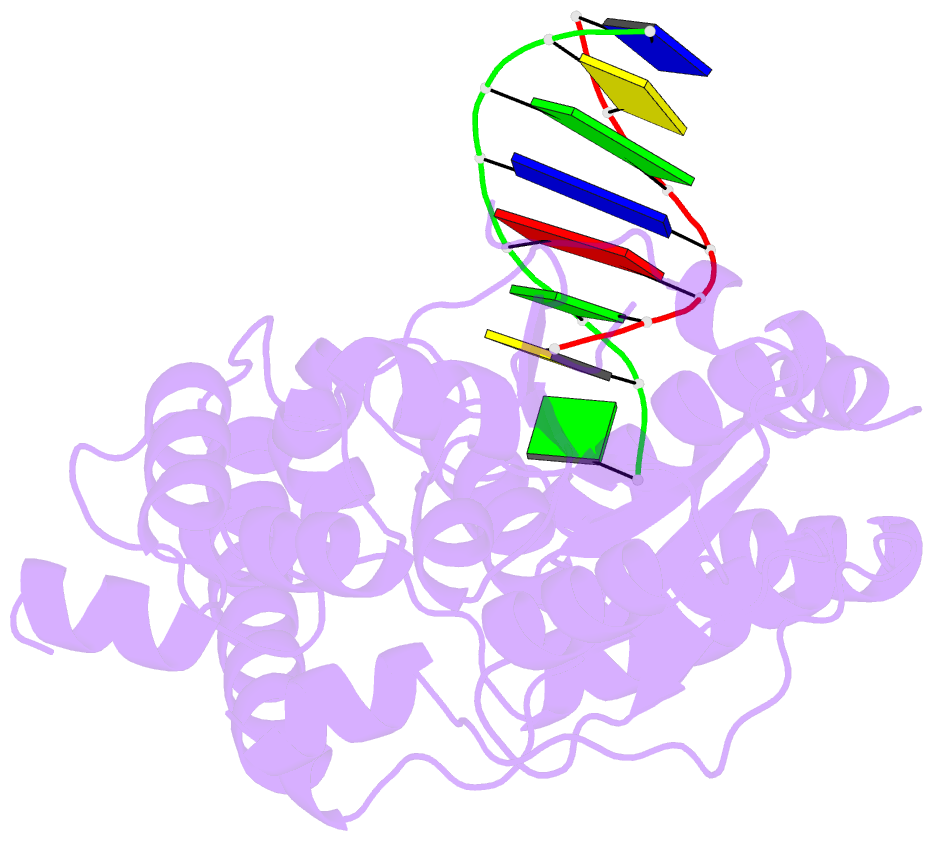
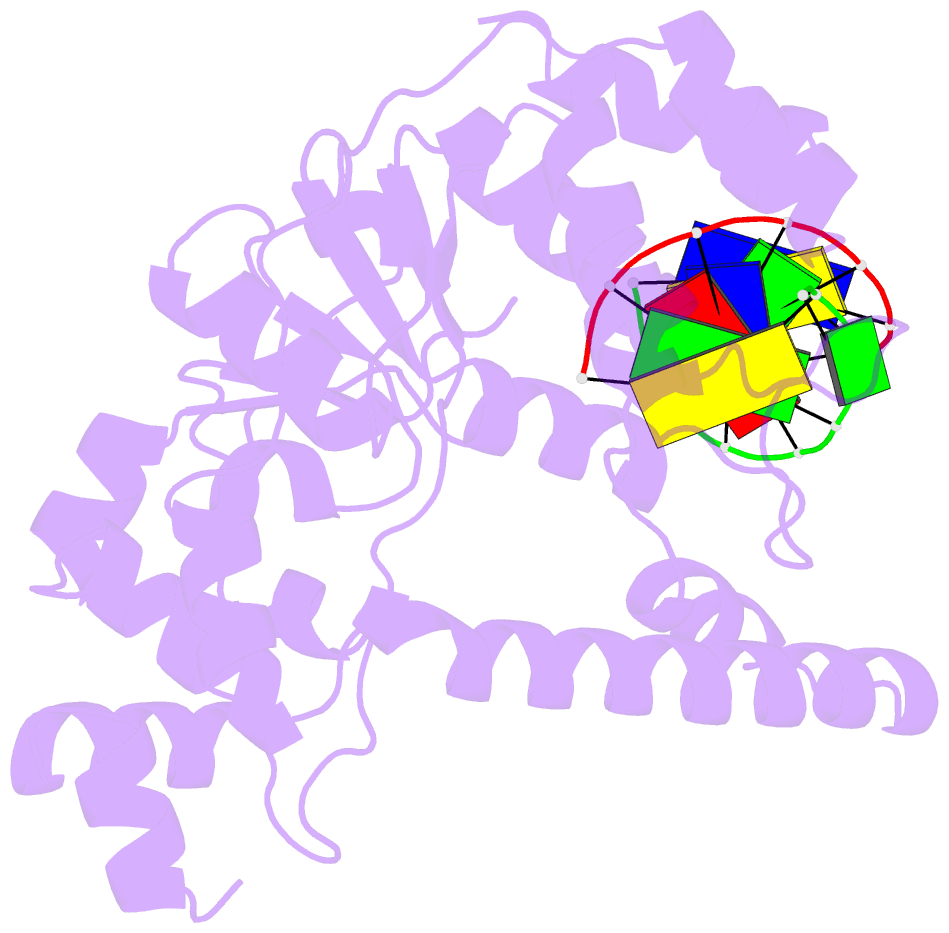
- Crystal structure of a. fulgidus fen-1 bound to DNA
- Reference
- Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA (2004): "Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair." Cell(Cambridge,Mass.), 116, 39-50. doi: 10.1016/S0092-8674(03)01036-5.
- Abstract
- Flap EndoNuclease-1 (FEN-1) and the processivity factor proliferating cell nuclear antigen (PCNA) are central to DNA replication and repair. To clarify the molecular basis of FEN-1 specificity and PCNA activation, we report here structures of FEN-1:DNA and PCNA:FEN-1-peptide complexes, along with fluorescence resonance energy transfer (FRET) and mutational results. FEN-1 binds the unpaired 3' DNA end (3' flap), opens and kinks the DNA, and promotes conformational closing of a flexible helical clamp to facilitate 5' cleavage specificity. Ordering of unstructured C-terminal regions in FEN-1 and PCNA creates an intermolecular beta sheet interface that directly links adjacent PCNA and DNA binding regions of FEN-1 and suggests how PCNA stimulates FEN-1 activity. The DNA and protein conformational changes, composite complex structures, FRET, and mutational results support enzyme-PCNA alignments and a kinked DNA pivot point that appear suitable to coordinate rotary handoffs of kinked DNA intermediates among enzymes localized by the three PCNA binding sites.