Summary information and primary citation
- PDB-id
- 1ry1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- translation
- Method
- cryo-EM (12.0 Å)
- Summary
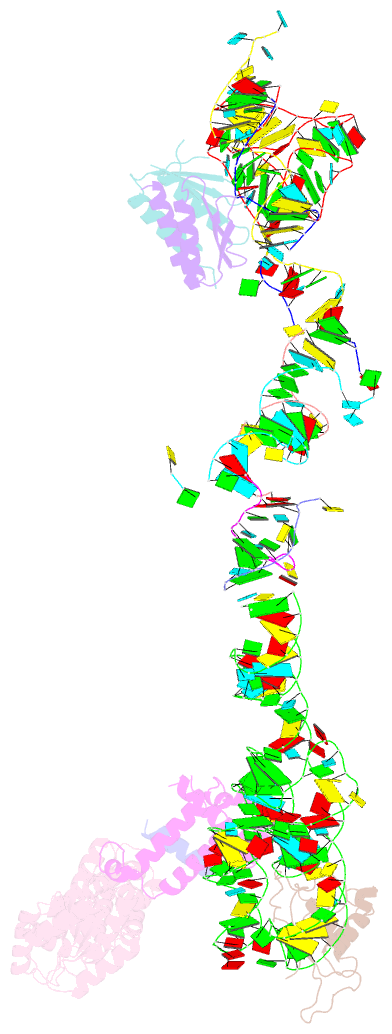
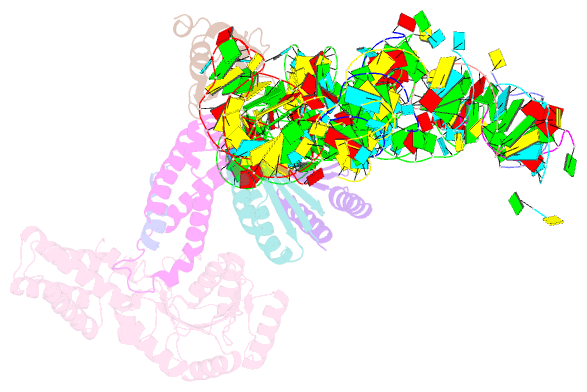
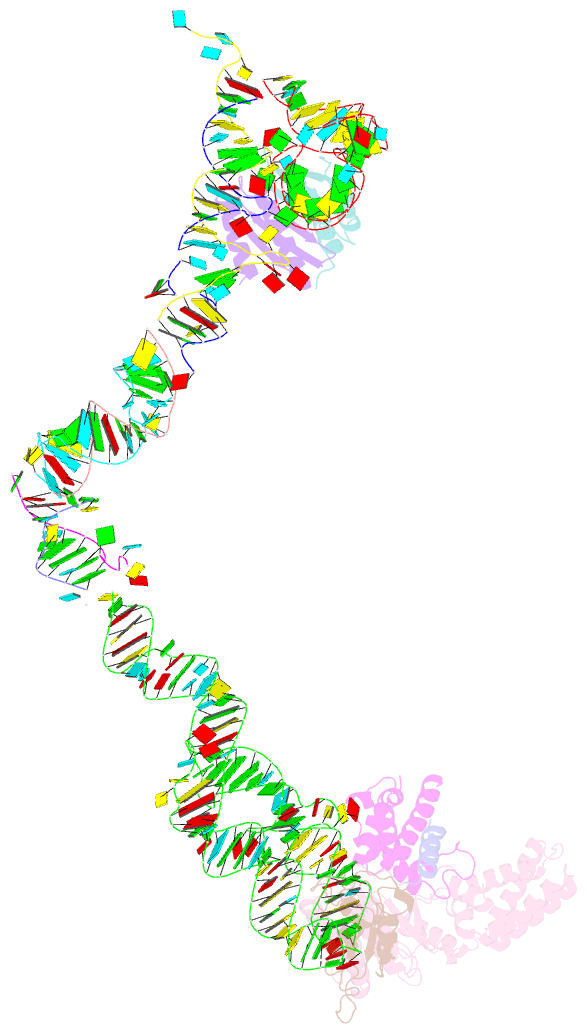
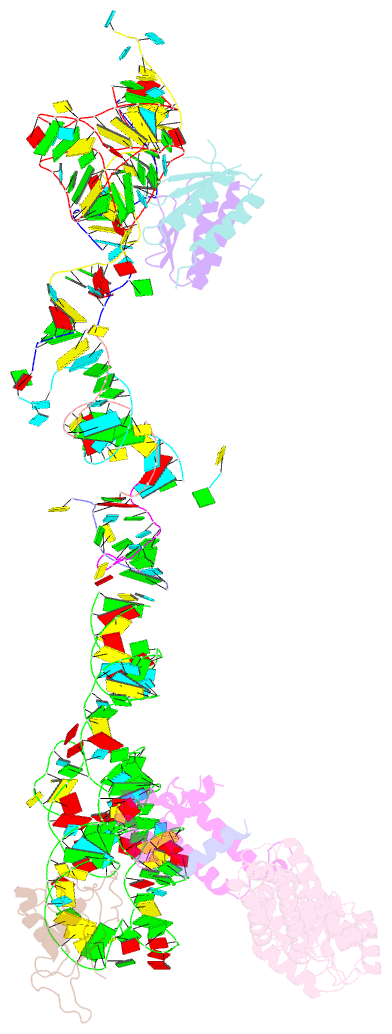
- Structure of the signal recognition particle interacting with the elongation-arrested ribosome
- Reference
- Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J, Beckmann R (2004): "Structure of the signal recognition particle interacting with the elongation-arrested ribosome." Nature, 427, 808-814. doi: 10.1038/nature02342.
- Abstract
- Cotranslational translocation of proteins across or into membranes is a vital process in all kingdoms of life. It requires that the translating ribosome be targeted to the membrane by the signal recognition particle (SRP), an evolutionarily conserved ribonucleoprotein particle. SRP recognizes signal sequences of nascent protein chains emerging from the ribosome. Subsequent binding of SRP leads to a pause in peptide elongation and to the ribosome docking to the membrane-bound SRP receptor. Here we present the structure of a targeting complex consisting of mammalian SRP bound to an active 80S ribosome carrying a signal sequence. This structure, solved to 12 A by cryo-electron microscopy, enables us to generate a molecular model of SRP in its functional conformation. The model shows how the S domain of SRP contacts the large ribosomal subunit at the nascent chain exit site to bind the signal sequence, and that the Alu domain reaches into the elongation-factor-binding site of the ribosome, explaining its elongation arrest activity.