Summary information and primary citation
- PDB-id
- 1s0v; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA-RNA hybrid
- Method
- X-ray (3.2 Å)
- Summary
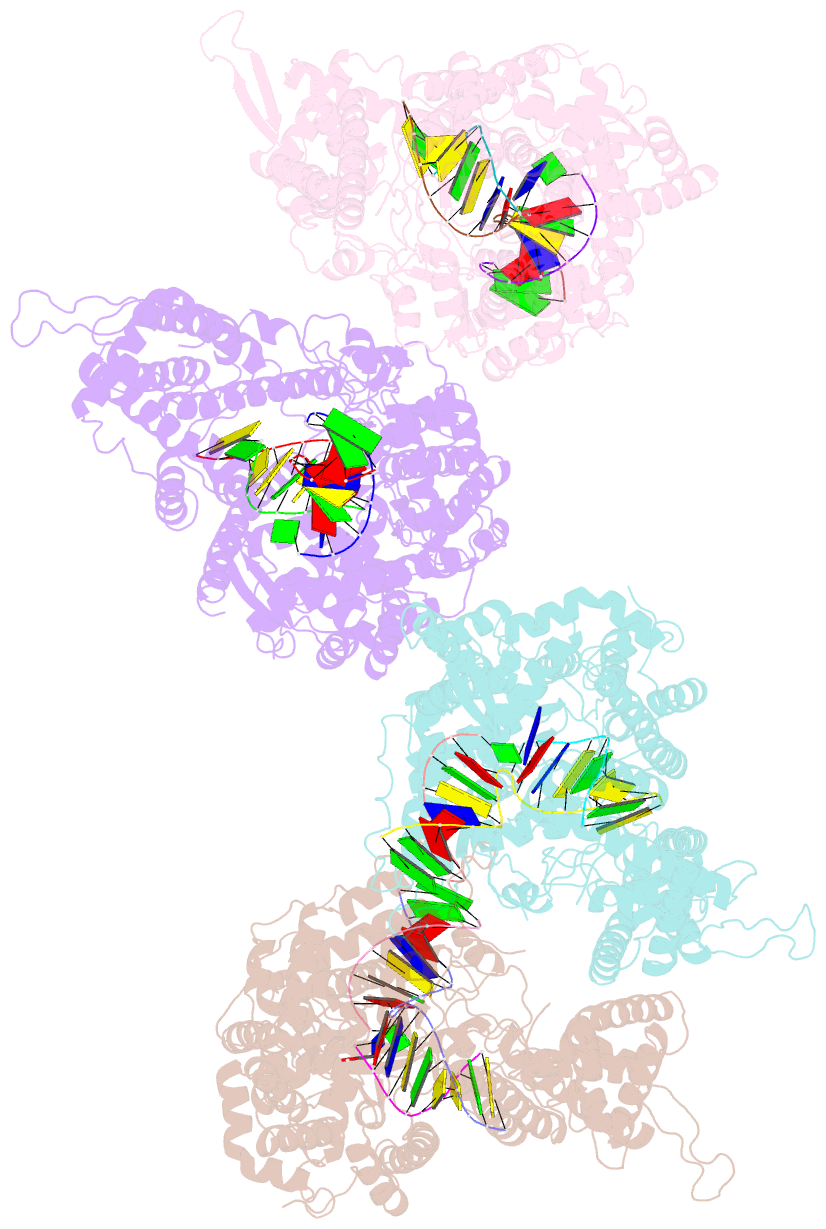
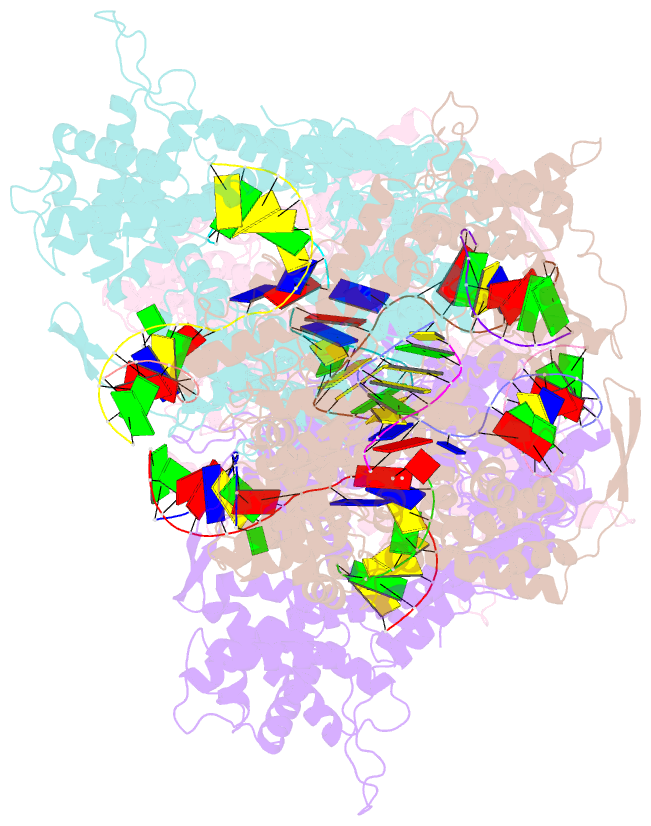
- Structural basis for substrate selection by t7 RNA polymerase
- Reference
- Temiakov D, Patlan V, Anikin M, McAllister WT, Yokoyama S, Vassylyev DG (2004): "Structural basis for substrate selection by t7 RNA polymerase." Cell(Cambridge,Mass.), 116, 381-391. doi: 10.1016/S0092-8674(04)00059-5.
- Abstract
- The mechanism by which nucleotide polymerases select the correct substrate is of fundamental importance to the fidelity of DNA replication and transcription. During the nucleotide addition cycle, pol I DNA polymerases undergo the transition from a catalytically inactive "open" to an active "closed" conformation. All known determinants of substrate selection are associated with the "closed" state. To elucidate if this mechanism is conserved in homologous single subunit RNA polymerases (RNAPs), we have determined the structure of T7 RNAP elongation complex with the incoming substrate analog. Surprisingly, the substrate specifically binds to RNAP in the "open" conformation, where it is base paired with the acceptor template base, while Tyr639 provides discrimination of ribose versus deoxyribose substrates. The structure therefore suggests a novel mechanism, in which the substrate selection occurs prior to the isomerization to the catalytically active conformation. Modeling of multisubunit RNAPs suggests that this mechanism might be universal for all RNAPs.