Summary information and primary citation
- PDB-id
- 1s32; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- structural protein-DNA
- Method
- X-ray (2.05 Å)
- Summary
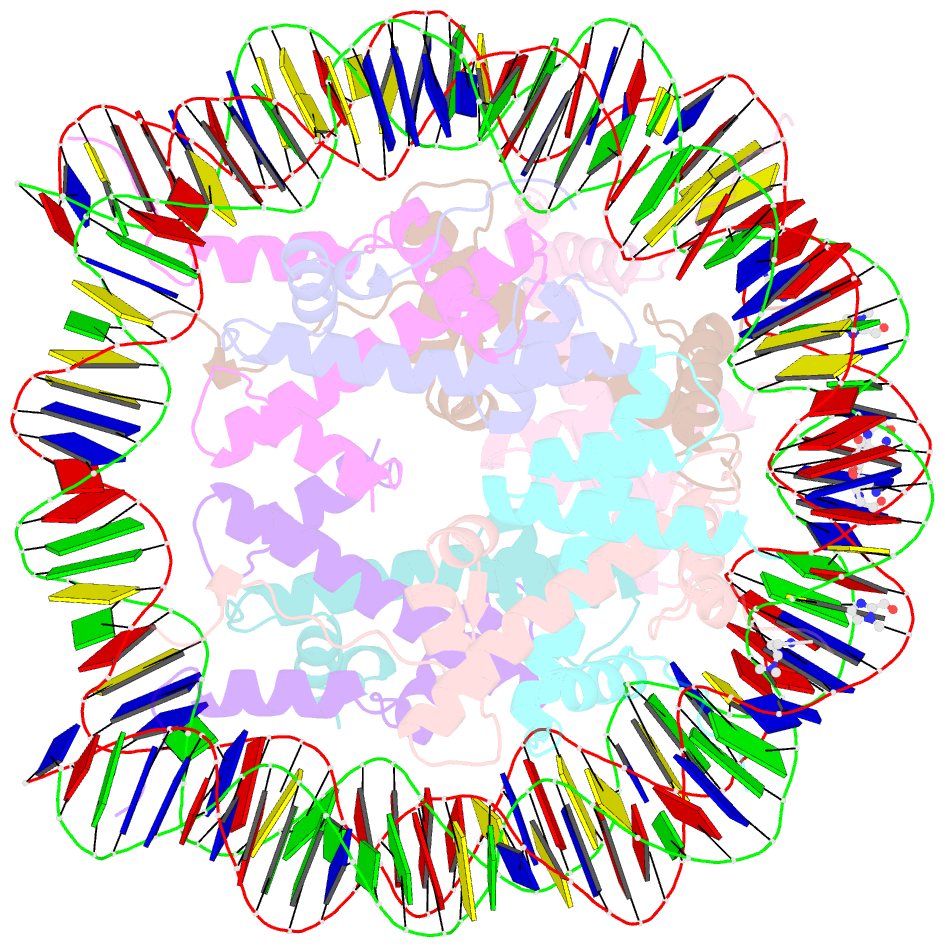
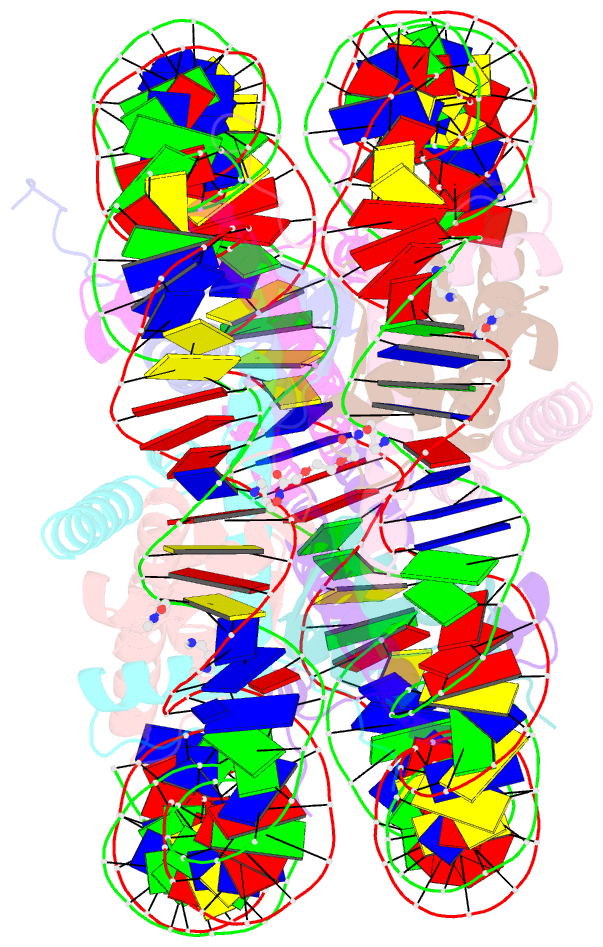
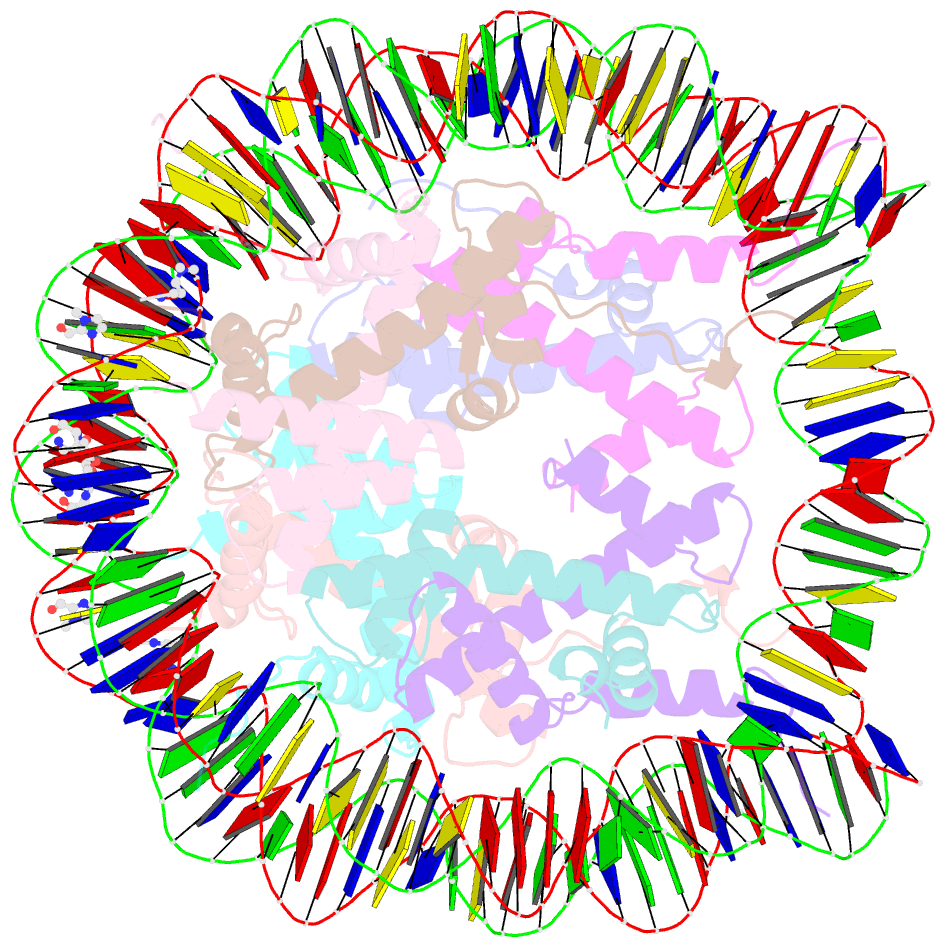
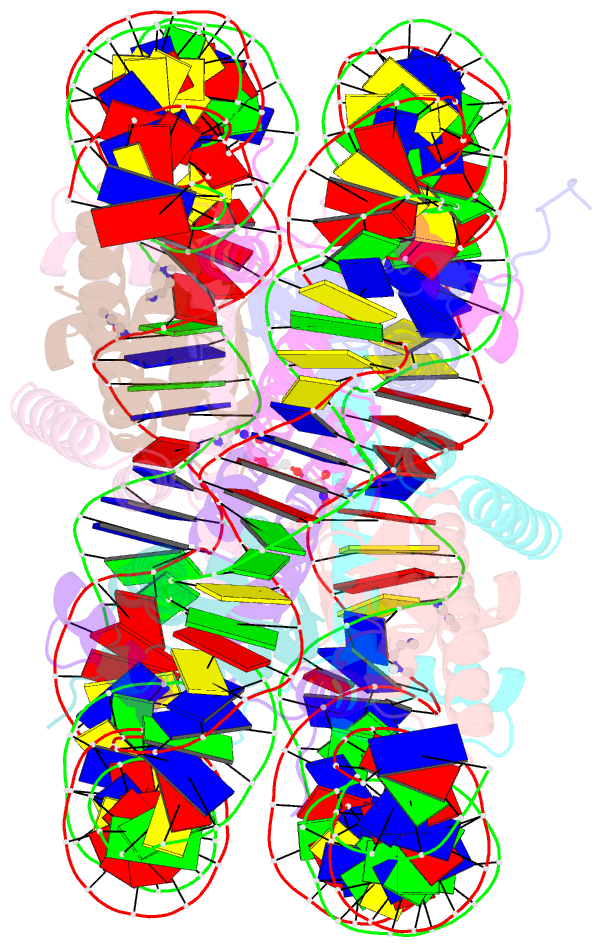
- Molecular recognition of the nucleosomal 'supergroove'
- Reference
- Edayathumangalam RS, Weyermann P, Gottesfeld JM, Dervan PB, Luger K (2004): "Molecular Recognition of the Nucleosomal 'Supergroove'." Proc.Natl.Acad.Sci.USA, 101, 6864-6869. doi: 10.1073/pnas.0401743101.
- Abstract
- Chromatin is the physiological substrate in all processes involving eukaryotic DNA. By organizing 147 base pairs of DNA into two tight superhelical coils, the nucleosome generates an architecture where DNA regions that are 80 base pairs apart on linear DNA are brought into close proximity, resulting in the formation of DNA "supergrooves." Here, we report the design of a hairpin polyamide dimer that targets one such supergroove. The 2-A crystal structure of the nucleosome-polyamide complex shows that the bivalent "clamp" effectively crosslinks the two gyres of the DNA superhelix, improves positioning of the DNA on the histone octamer, and stabilizes the nucleosome against dissociation. Our findings identify nucleosomal supergrooves as platforms for molecular recognition of condensed eukaryotic DNA. In vivo, supergrooves may foster synergistic protein-protein interactions by bringing two regulatory elements into juxtaposition. Because supergroove formation is independent of the translational position of the DNA on the histone octamer, accurate nucleosome positioning over regulatory elements is not required for supergroove participation in eukaryotic gene regulation.