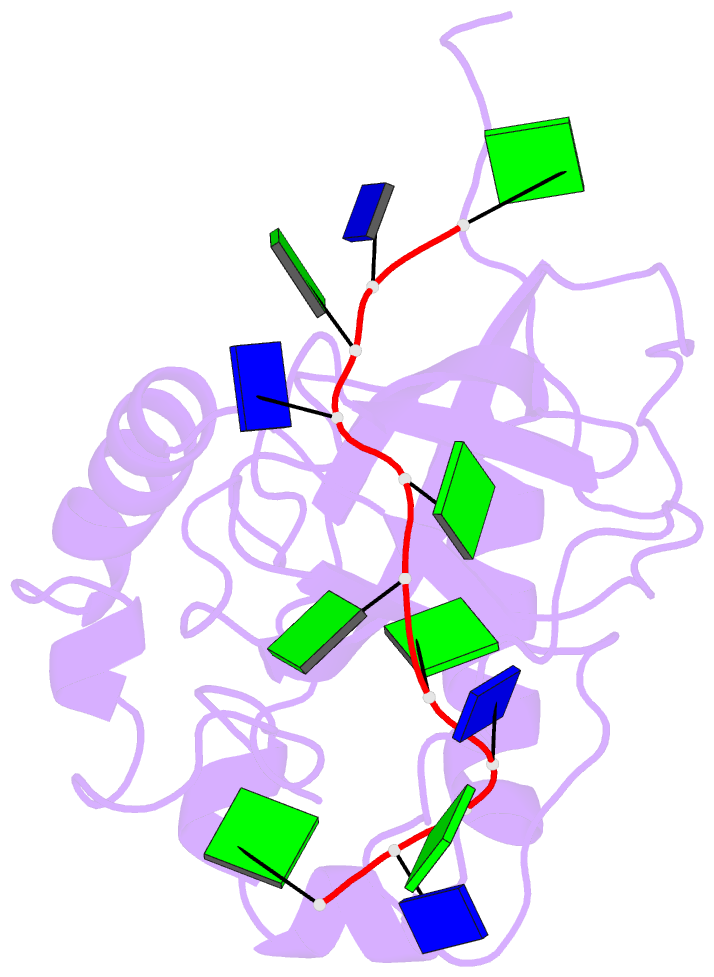
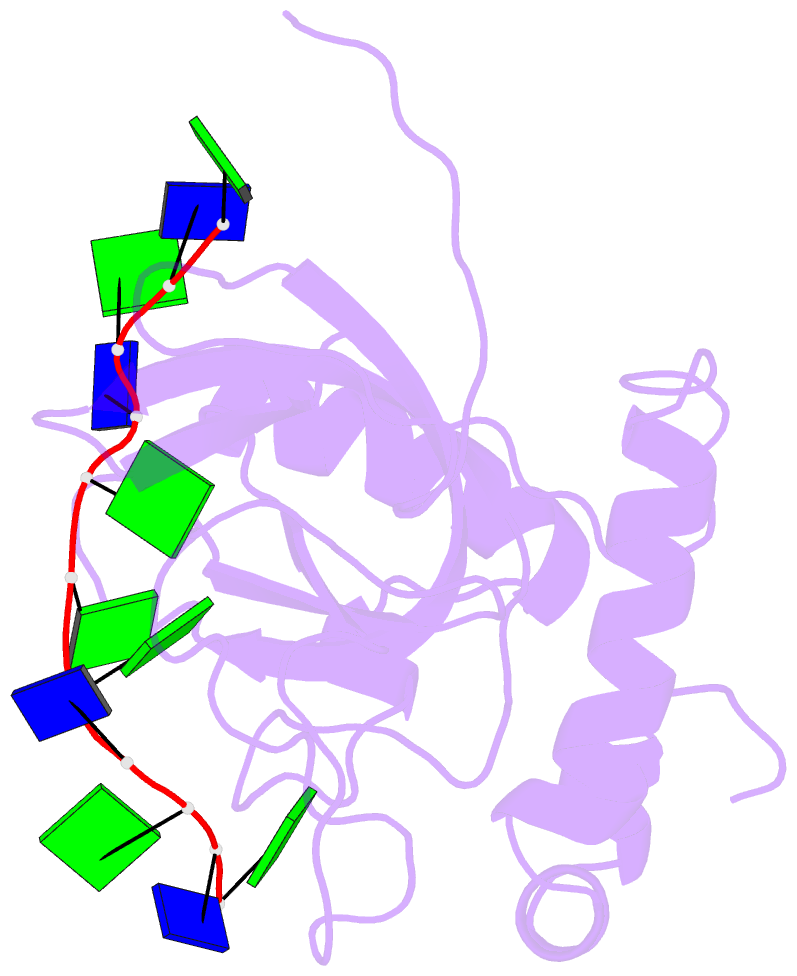
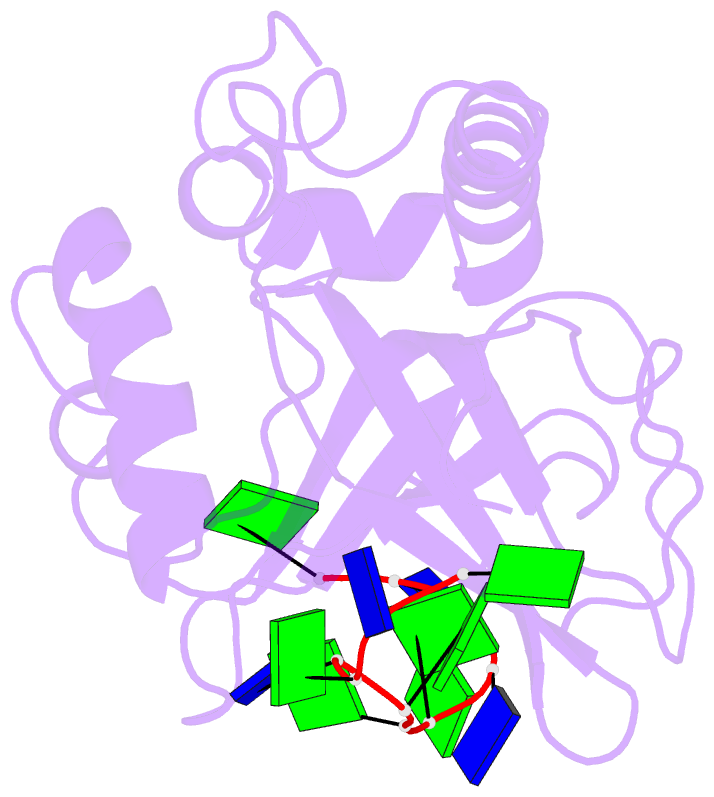
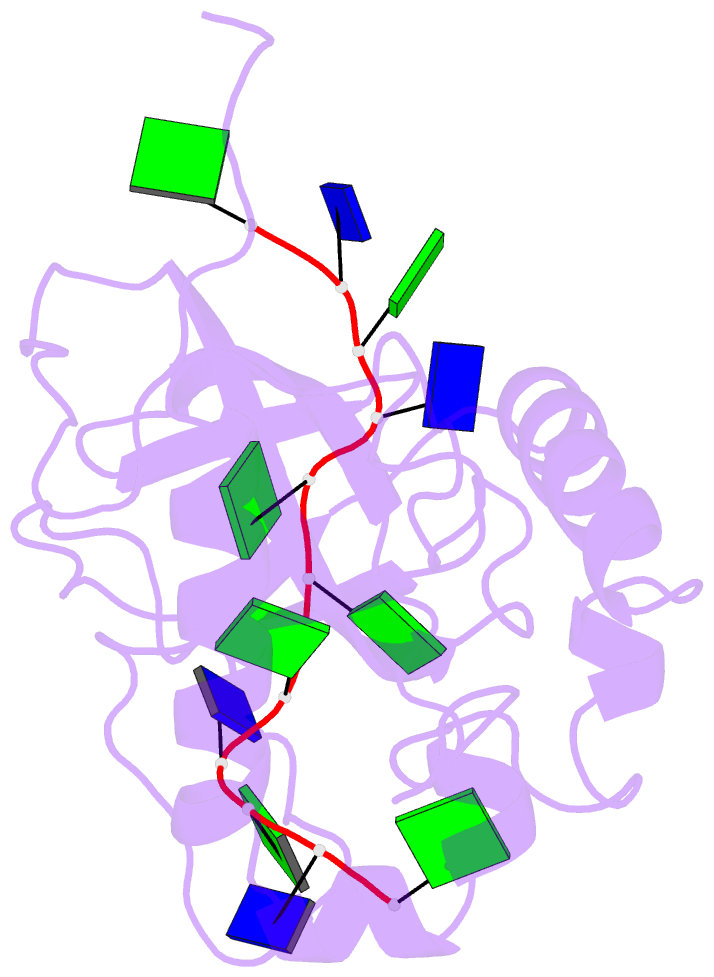
Summary information and primary citation
- PDB-id
- 1s40; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- cell cycle-DNA
- Method
- NMR
- Summary
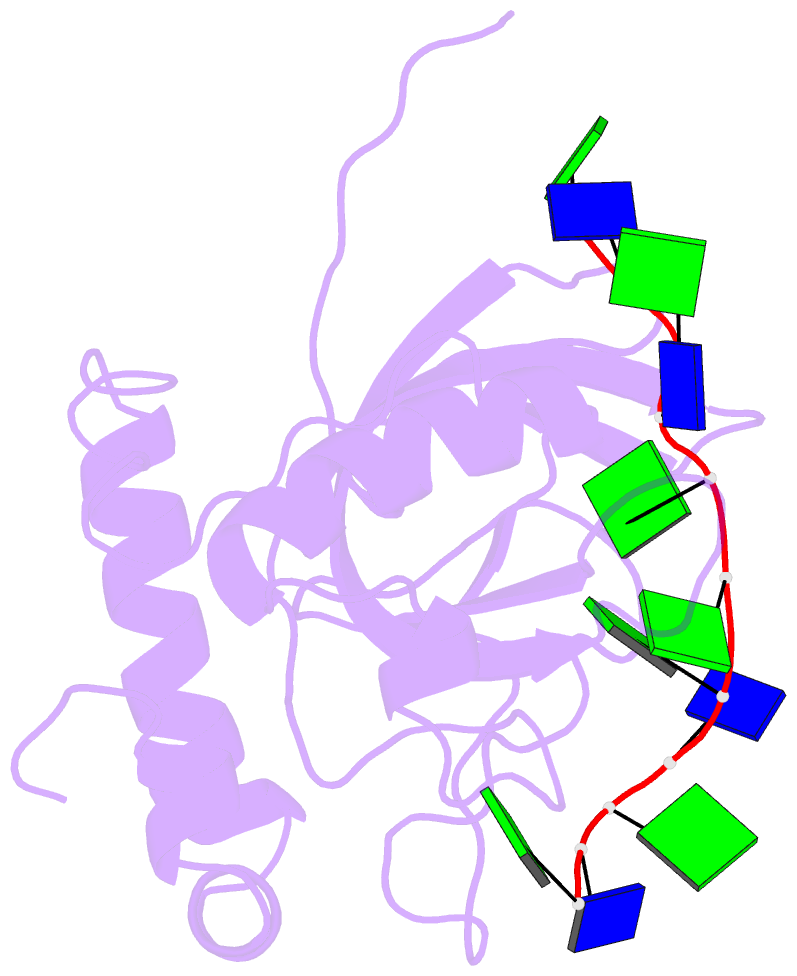
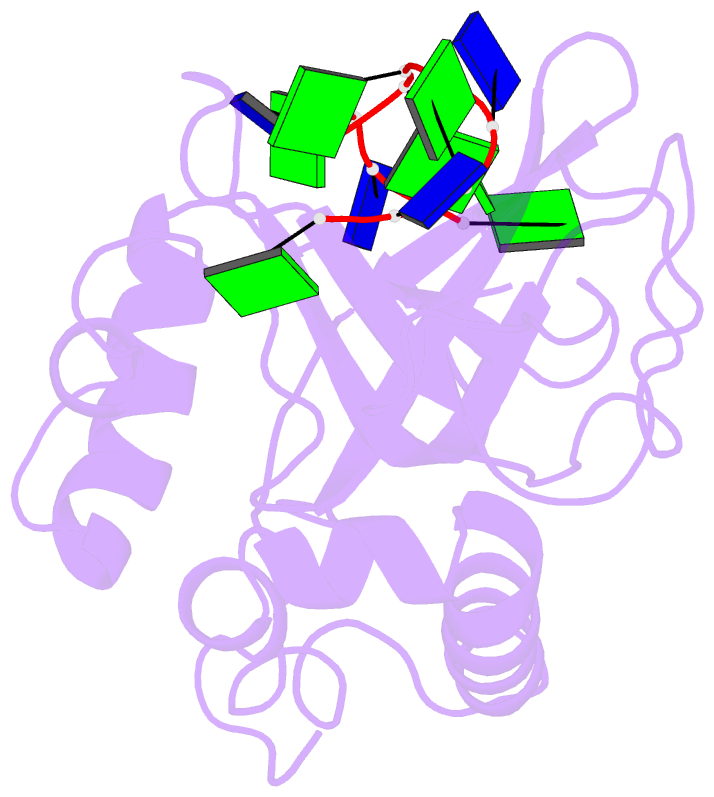
- Solution structure of the cdc13 DNA-binding domain complexed with a single-stranded telomeric DNA 11-mer
- Reference
- Mitton-Fry RM, Anderson EM, Theobald DL, Glustrom LW, Wuttke DS (2004): "Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13." J.Mol.Biol., 338, 241-255. doi: 10.1016/j.jmb.2004.01.063.
- Abstract
- The essential budding yeast telomere-binding protein Cdc13 is required for telomere replication and end protection. Cdc13 specifically binds telomeric, single-stranded DNA (ssDNA) 3' overhangs with high affinity using an OB-fold domain. We have determined the high-resolution solution structure of the Cdc13 DNA-binding domain (DBD) complexed with a cognate telomeric ssDNA. The ssDNA wraps around one entire face of the Cdc13-DBD OB-fold in an extended, irregular conformation. Recognition of the ssDNA bases occurs primarily through aromatic, basic, and hydrophobic amino acid residues, the majority of which are evolutionarily conserved among budding yeast species and contribute significantly to the energetics of binding. Contacting five of 11 ssDNA nucleotides, the large, ordered beta2-beta3 loop is crucial for complex formation and is a unique elaboration on the binding mode commonly observed in OB-fold proteins. The sequence-specific Cdc13-DBD/ssDNA complex presents a complementary counterpoint to the interactions observed in the Oxytricha nova telomere end-binding and Schizosaccharomyces pombe Pot1 complexes. Analysis of the Cdc13-DBD/ssDNA complex indicates that molecular recognition of extended single-stranded nucleic acids may proceed via a folding-type mechanism rather than resulting from specific patterns of hydrogen bonds. The structure reported here provides a foundation for understanding the mechanism by which Cdc13 recognizes GT-rich heterogeneous sequences with both unusually strong affinity and high specificity.